Displayed Output
The following output is produced by the GENMOD procedure.
Note that some of the tables are optional and appear only in
conjunction with the REPEATED statement and its options or with
options in the MODEL statement. For details, see
the section "ODS Table Names".
Model Information
PROC GENMOD displays the following model information:
- data set name
- response distribution
- link function
- response variable name
- offset variable name
- frequency variable name
- scale weight variable name
- number of observations used
- number of events if events/trials
format is used for response
- number of trials if events/trials
format is used for response
- sum of frequency weights
- number of missing values in data set
- number of invalid observations (for example, negative
or 0 response values with gamma distribution or
number of observations with events greater than
trials with binomial distribution)
Class Level Information
If you use classification variables in the model, PROC
GENMOD displays the levels of classification variables
specified in the CLASS statement and in the MODEL statement.
The levels are displayed in the same sorted order
used to generate columns in the design matrix.
If you specify an ordinal model for the multinomial distribution,
a table titled "Response Profile" is displayed containing
the ordered values of the response variable and the
number of occurrences of the values used in the model.
Iteration History for Parameter Estimates
If you specify the ITPRINT model option, PROC GENMOD
displays
a table containing the following for each iteration
in the Newton-Raphson procedure for model fitting:
- iteration number
- ridge value
- log likelihood
- values of all parameters in the model
Criteria for Assessing Goodness of Fit
PROC GENMOD displays the following criteria for assessing goodness of fit:
- degrees of freedom for deviance and Pearson's chi-square,
equal to the number of observations minus the number of
regression parameters estimated
- deviance
- deviance divided by degrees of freedom
- scaled deviance
- scaled deviance divided by degrees of freedom
- Pearson's chi-square
- Pearson's chi-square divided by degrees of freedom
- scaled Pearson's chi-square
- scaled Pearson's chi-square divided by degrees of freedom
- log likelihood
Last Evaluation of the Gradient
If you specify the model option ITPRINT, the GENMOD procedure
displays the last evaluation of the gradient vector.
Last Evaluation of the Hessian
If you specify the model option ITPRINT, the GENMOD procedure
displays the last evaluation of the Hessian matrix.
Analysis of (Initial) Parameter Estimates
The "Analysis of (Initial) Parameter Estimates"
table contains the results from fitting
a generalized linear model to the data.
If you specify the REPEATED statement, these GLM parameter
estimates are used as initial values for the GEE solution.
For each parameter in the model, PROC GENMOD displays the following:
- the parameter name
- -
- the variable name for continuous
regression variables
- -
- the variable name and level for
classification variables and interactions
involving classification variables
- -
- SCALE for the scale variable related to
the dispersion parameter
- degrees of freedom for the parameter
- estimate value
- standard error
- Wald chi-square value
- p-value based on the chi-square distribution
- confidence limits (Wald or profile likelihood) for
parameters
Estimated Covariance Matrix
If you specify the model option COVB, the GENMOD procedure
displays the estimated covariance matrix, defined as the
inverse of the information matrix at the final iteration.
This is based on the expected information matrix if the
EXPECTED option is specified in the MODEL statement.
Otherwise, it is based on the Hessian
matrix used at the final iteration.
This is, by default, the observed Hessian unless
altered by the SCORING option in the MODEL statement.
Estimated Correlation Matrix
If you specify the CORRB model option, PROC
GENMOD displays the estimated correlation matrix.
This is based on the expected information matrix if the
EXPECTED option is specified in the MODEL statement.
Otherwise, it is based on the Hessian
matrix used at the final iteration.
This is, by default, the observed Hessian unless
altered by the SCORING option in the MODEL statement.
Iteration History for LR Confidence Intervals
If you specify the ITPRINT and LRCI model options,
PROC GENMOD displays an iteration history table for profile
likelihood-based confidence intervals.
For each parameter in the model, PROC GENMOD displays the following:
- parameter identification number
- iteration number
- log likelihood value
- parameter values
Likelihood Ratio-Based Confidence Intervals for Parameters
If you specify the LRCI and the ITPRINT options in the
MODEL statement, a table
is displayed summarizing profile likelihood-based
confidence intervals for all parameters.
The table contains the following for each parameter in the model:
- confidence coefficient
- parameter identification number
- lower and upper endpoints of confidence
intervals for the parameter
- values of all other parameters at the solution
LR Statistics for Type 1 Analysis
If you specify the TYPE1 model option, a table containing
the following is displayed for each effect in the model:
If you specify either the SCALE=DEVIANCE or SCALE=PEARSON option in the
MODEL statement, columns containing the following are displayed:
- name of effect
- deviance for the model including the
effect and all previous effects
- numerator degrees of freedom
- denominator degrees of freedom
- chi-square statistic for testing
the significance of the effect
- p-value computed from the chi-square
distribution with numerator degrees of freedom
- F statistic for testing the significance of the effect
- p-value based on the F distribution
Iteration History for Type 3 Contrasts
If you specify the model options ITPRINT and TYPE3,
an iteration history table is displayed for fitting the
model with Type 3 contrast constraints for each effect.
The table contains the following:
- effect name
- iteration number
- ridge value
- log likelihood
- values of all parameters
LR Statistics for Type 3 Analysis
If you specify the TYPE3 model option, a table containing
the following is displayed for each effect in the model:
- name of the effect
- likelihood ratio statistic for testing
the significance of the effect
- degrees of freedom for effect
- p-value computed from the chi-square distribution
If you specify either the SCALE=DEVIANCE or SCALE=PEARSON option in the
MODEL statement, columns containing the following are displayed:
- name of the effect
- likelihood ratio statistic for testing
the significance of the effect
- F statistic for testing the significance of the effect
- numerator degrees of freedom
- denominator degrees of freedom
- p-value based on the F distribution
- p-value computed from the chi-square
distribution with numerator degrees of freedom
Wald Statistics for Type 3 Analysis
If you specify the TYPE3 and WALD model options, a table
containing the following is displayed for each effect in the model:
- name of effect
- degrees of freedom for effect
- Wald statistic for testing the significance of the effect
- p-value computed from the chi-square distribution
Parameter Information
If you specify the ITPRINT, COVB, CORRB,
WALDCI, or LRCI option
in the MODEL statement, or if you specify a CONTRAST statement,
a table is displayed that identifies parameters with
numbers, rather than names, for use in tables and matrices
where a compact identifier for parameters is helpful.
For each parameter, the table contains the following:
- a number that identifies the parameter
- the parameter name, including level information
for effects containing classification variables
Observation Statistics
If you specify the OBSTATS option in the MODEL statement,
PROC GENMOD displays a table containing miscellaneous statistics.
For each observation in the input
data set, the following are displayed:
- the value of the response variable,
denoted by the variable name
- the predicted value of the mean, denoted by PRED
- the value of the linear predictor,
denoted by XBETA.
The value of an OFFSET
variable is not added to the linear predictor.
- the estimated standard error of the linear predictor,
denoted by STD
- the value of the negative of the weight in the Hessian
matrix at the final iteration, denoted by HESSWGT.
This is the expected weight if the EXPECTED option
is specified in the MODEL statement.
Otherwise, it is the weight used in the final iteration.
That is, it is the observed weight unless
the SCORING= option has been specified.
- approximate lower and upper endpoints for a confidence
interval for the predicted value of the mean, denoted
by LOWER and UPPER
- raw residual, denoted by RESRAW
- Pearson residual, denoted by RESCHI
- deviance residual, denoted by RESDEV
- standardized Pearson residual, denoted by STDRESCHI
- standardized deviance residual, denoted by STDRESDEV
- likelihood residual, denoted by RESLIK
ESTIMATE Statement Results
If you specify a REPEATED statement, the ESTIMATE statement results
apply to the specified GEE model. Otherwise, they apply to
the specified generalized linear model.
The following are displayed for each ESTIMATE statement:
- contrast label
- estimated value of the contrast
- standard error of the estimate
- significance level
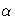
-
 confidence intervals for contrast
confidence intervals for contrast
- Wald chi-square statistic for the contrast
- p-value computed from the chi-square distribution
If you specify the EXP option, an additional row is displayed
with statistics for the exponentiated value of the contrast.
CONTRAST Coefficients
If you specify the CONTRAST or ESTIMATE statement and you specify the E option,
a table titled "Coefficients For
Contrast label" is displayed, where label
is the label specified in the CONTRAST statement.
The table contains the following:
- the contrast label
- the rows of the contrast matrix
Iteration History for Contrasts
If you specify the ITPRINT option, an iteration history table is displayed
for fitting the model with contrast constraints for each effect.
The table contains the following for each
contrast defined in a CONTRAST statement:
- contrast label
- iteration number
- ridge value
- log likelihood
- values of all parameters
CONTRAST Statement Results
If you specify a REPEATED statement, the CONTRAST statement results
apply to the specified GEE model. Otherwise, they apply to
the specified generalized linear model.
The following are displayed for each CONTRAST statement:
- contrast label
- degrees of freedom for the contrast
- likelihood ratio, score, or Wald statistic for testing
the significance of the contrast. Score statistics are
used in GEE models, likelihood ratio statistics are used in
generalized linear models, and Wald statistics are used in both.
- p-value computed from the chi-square distribution
- the type of statistic computed for this contrast: Wald, LR, or score
If you specify either the SCALE=DEVIANCE or SCALE=PEARSON option for generalized
linear models,
columns containing the following are displayed:
- contrast label
- likelihood ratio statistic for testing the significance
of the contrast
- F statistic for testing the significance of the contrast
- numerator degrees of freedom
- denominator degrees of freedom
- p-value based on the F distribution
- p-value computed from the chi-square distribution with
numerator degrees of freedom
LSMEANS Coefficients
If you specify the LSMEANS statement and you specify the E option,
a table titled
"Coefficients for effect Least Squares Means" is displayed, where effect
is the effect specified in the LSMEANS statement.
The table contains the following:
- the effect names
- the rows of least squares means coefficients
Least Squares Means
If you specify the LSMEANS statement
a table titled
"Least Squares Means" is displayed.
The table contains the following:
- the effect names
- for each level of each effect,
- -
- the least squares mean estimate
- -
- standard error
- -
- chi-square value
- -
- p-value computed from the chi-square distribution
If you specify the DIFF option, a table titled
"Differences of Least Squares Means" is displayed
containing corresponding statistics for the differences between
the least squares means for the levels of each effect.
GEE Model Information
If you specify the REPEATED statement, the following are displayed:
- correlation structure of the working correlation matrix
or the log odds ratio structure
- within-subject effect
- subject effect
- number of clusters
- correlation matrix dimension
- minimum and maximum cluster size
Log Odds Ratio Parameter Information
If you specify the REPEATED statement and specify a log odds ratio
model for binary data with the LOGOR= option, then a table is
displayed showing the correspondence between data pairs and log odds
ratio model parameters.
Iteration History for GEE Parameter Estimates
If you specify the REPEATED statement and the MODEL
statement option ITPRINT, an iteration
history table for GEE parameter estimates is displayed.
The table contains the following:
- parameter identification number
- iteration number
- values of all parameters
Last Evaluation of the Generalized Gradient and Hessian
If you specify the REPEATED statement and select ITPRINT
as a model option, PROC GENMOD displays
the last generalized gradient and Hessian matrix
in the GEE iterative parameter estimation process.
GEE Parameter Estimate Covariance Matrices
If you specify the REPEATED statement and the COVB option,
PROC GENMOD displays both model-based and empirical parameter
estimate covariance matrices.
GEE Parameter Estimate Correlation Matrices
If you specify the REPEATED statement and the CORRB option,
PROC GENMOD displays both model-based and empirical parameter
estimate covariance matrices.
GEE Working Correlation Matrix
If you specify the REPEATED statement and the CORRW option,
PROC GENMOD displays the exchangeable working correlation matrix.
Analysis of GEE Parameter Estimates
If you specify the REPEATED statement, PROC GENMOD
uses empirical standard error estimates to compute and
display the following for each parameter in the model:
- the parameter name
- -
- the variable name for continuous
regression variables
- -
- the variable name and level for classification
variables and interactions involving
classification variables
- -
- "Scale" for the scale variable
related to the dispersion parameter
- parameter estimate
- standard error
- 95% confidence interval
- Z score and p-value
If you specify the MODELSE option in the REPEATED statement,
a table based on model-based standard errors is also produced.
GEE Observation Statistics
If you specify the OBSTATS option in the REPEATED statement,
PROC GENMOD displays a table containing miscellaneous statistics.
For each observation in the input
data set, the following are displayed:
- the value of the response variable and all other
variables in the model, denoted by the variable names
- the predicted value of the mean, denoted by PRED
- the value of the linear predictor, denoted by XBETA
- the standard error of the linear predictor, denoted by STD
- confidence limits for the predicted values,
denoted by LOWER and UPPER
- raw residual, denoted by RESRAW
- Pearson residual, denoted by RESCHI
Copyright © 1999 by SAS Institute Inc., Cary, NC, USA. All rights reserved.