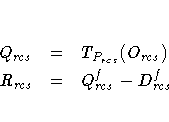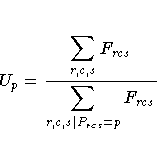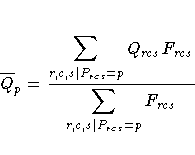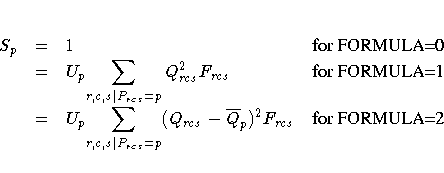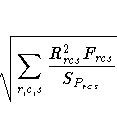Formulas
The following notation is used:
- Ap
-
intercept for partition p
- Bp
- slope for partition p
- Cp
- power for partition p
- Drcs
- distance computed from the model between objects r and c for
subject s
- Frcs
-
data weight for objects r and c for subject s
obtained from the cth WEIGHT variable, or 1 if there is no WEIGHT
statement
- f
- value of the FIT= option
- N
- number of objects
- Orcs
- observed dissimilarity between objects r and c for subject s
- Prcs
- partition index for objects r and c for subject s
- Qrcs
- dissimilarity after applying any applicable estimated transformation for
objects r and c for subject s
- Rrcs
-
residual for objects r and c for subject s
- Sp
- standardization factor for partition p
- Tp(·)
- estimated transformation for partition p
- Vsd
- coefficient for subject s on dimension d
- Xnd
- coordinate for object n on dimension d
Summations are taken over nonmissing values.
Distances are computed from the model as
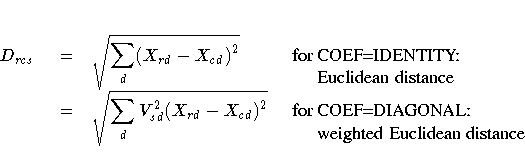
Partition indexes are
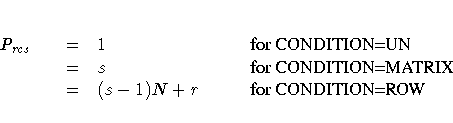
The estimated transformation for each partition is
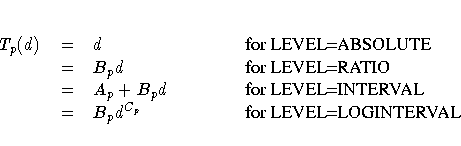
For LEVEL=ORDINAL, Tp(·) is computed as a
least-squares monotone transformation.
For LEVEL=ABSOLUTE, RATIO, or INTERVAL, the residuals are computed as
![Q_{rcs} &=& O_{rcs} \R_{rcs} &=& Q_{rcs}^f - [T_{P_{rcs}}(D_{rcs})]^f](images/mdseq11.gif)
For LEVEL=ORDINAL, the residuals are computed as
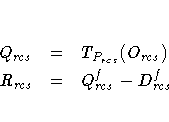
If f is 0, then natural logarithms are used in place of
the fth
powers.
For each partition, let
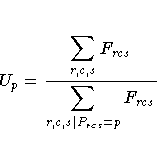
and
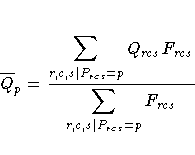
Then the standardization factor for each partition is
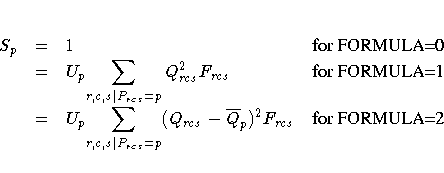
The badness-of-fit criterion that the MDS procedure tries to
minimize is
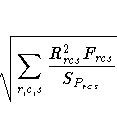
Copyright © 1999 by SAS Institute Inc., Cary, NC, USA. All rights reserved.
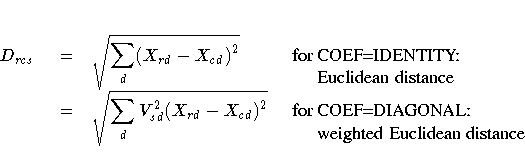
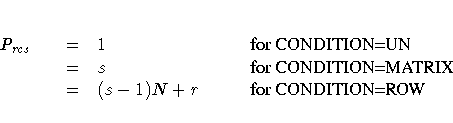
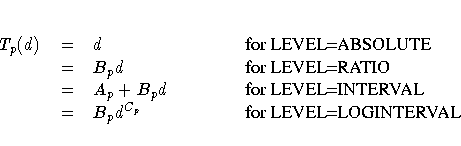
![Q_{rcs} &=& O_{rcs} \R_{rcs} &=& Q_{rcs}^f - [T_{P_{rcs}}(D_{rcs})]^f](images/mdseq11.gif)