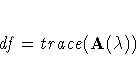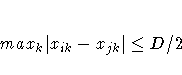MODEL Statement
- MODEL dependents = < regression variables >
(smoothing variables) < /options > ;
The MODEL statement specifies the dependent variables, the independent
regression variables, which are listed with no parentheses, and the
independent smoothing variables, which are listed inside parentheses.
The regression variables are optional. At least one smoothing variable
is required, and it must be listed after the regression variables. No
variables can be listed in both the regression variable list and the
smoothing variable list.
If you specify more than one dependent variable, PROC TPSPLINE
calculates a thin-plate smoothing spline estimate for each dependent
variable, using the regression variables and smoothing variables
specified on the right-hand side.
If you specify regression variables, PROC TPSPLINE fits a
semiparametric model using the regression variables as the linear part
of the model.
You can specify the following options in the MODEL statement.
- ALPHA=number
-
specifies the significance level
 of the confidence limits on
the final thin-plate smoothing spline estimate when you request
confidence limits to be included in the output data set. Specify
number as a value between 0 and 1. The default value is 0.05.
See
the "OUTPUT Statement" section for more information on the OUTPUT statement.
of the confidence limits on
the final thin-plate smoothing spline estimate when you request
confidence limits to be included in the output data set. Specify
number as a value between 0 and 1. The default value is 0.05.
See
the "OUTPUT Statement" section for more information on the OUTPUT statement.
- DF=number
-
specifies the degrees of freedom of the thin-plate smoothing
spline estimate, defined as
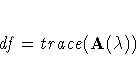
where  is the hat matrix.
Specify number as a value between zero and the number of
unique design points.
is the hat matrix.
Specify number as a value between zero and the number of
unique design points.
- DISTANCE=number
- D=number
-
defines a range such that if two data points
(xi, zi) and (xj, zj)
satisfy
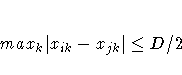
then these data points are treated as
replicates, where xi are the smoothing variables and zi
are the regression variables.
You can use the DISTANCE= option to reduce the number of unique
design points by treating nearby data as
replicates. This can be useful when you have a
large data set. The default value is 0.
- LAMBDA0=number
-
specifies the smoothing parameter,
 , to be used in the
thin-plate smoothing spline estimate.
By default, PROC TPSPLINE uses the
, to be used in the
thin-plate smoothing spline estimate.
By default, PROC TPSPLINE uses the  parameter
that minimizes the GCV function for the final fit.
The LAMBDA0= value must be positive.
parameter
that minimizes the GCV function for the final fit.
The LAMBDA0= value must be positive.
- LAMBDA=list-of-values
-
specifies a set of values for the
 parameter.
PROC TPSPLINE returns a GCV
value for each
parameter.
PROC TPSPLINE returns a GCV
value for each  point that you specify. You can use the LAMBDA=
option to study the GCV function curve for a set of values for
point that you specify. You can use the LAMBDA=
option to study the GCV function curve for a set of values for
 . All values listed in the LAMBDA= option must be positive.
. All values listed in the LAMBDA= option must be positive.
- LOGNLAMBDA0=number
- LOGNL0=number
-
specifies the smoothing parameter
 on the
on the  scale.
If you specify both the LOGNL0= and LAMBDA0= options, only the value
provided by the LOGNL0= option is used. By default,
PROC TPSPLINE uses the
scale.
If you specify both the LOGNL0= and LAMBDA0= options, only the value
provided by the LOGNL0= option is used. By default,
PROC TPSPLINE uses the  parameter
that minimizes the GCV function for the estimate.
parameter
that minimizes the GCV function for the estimate.
- LOGNLAMBDA=list-of-values
- LOGNL=list-of-values
-
specifies a set of values for the
 parameter
on the
parameter
on the  scale.
PROC TPSPLINE returns a
GCV value for each
scale.
PROC TPSPLINE returns a
GCV value for each  point that you specify.
You can use the LOGNLAMBDA= option to study the GCV function curve for a
set of
point that you specify.
You can use the LOGNLAMBDA= option to study the GCV function curve for a
set of  values. If you specify both the LOGNL= and
LAMBDA= options, only the list of values provided by LOGNL= option is
used.
values. If you specify both the LOGNL= and
LAMBDA= options, only the list of values provided by LOGNL= option is
used.
In some cases, the LOGNL= option may be prefered over the LAMBDA=
option. Because the LAMBDA= value must be positive, a small change in
that value can result in a major change in the GCV value. If you
instead specify  on the log10 scale, the allowable range is
enlarged to include negative values. Thus, the GCV function is less
sensitive to changes in LOGNLAMBDA.
on the log10 scale, the allowable range is
enlarged to include negative values. Thus, the GCV function is less
sensitive to changes in LOGNLAMBDA.
- M=number
-
specifies the order of the derivative
in the penalty term. The M= value must be a positive
integer. The default value is the max(2, INT(d/2)+1), where
d is the number of smoothing variables.
Copyright © 1999 by SAS Institute Inc., Cary, NC, USA. All rights reserved.