Part A
From text page 254-255, 6.15 c, d, e, f, g, 6.16, 6.17. Page 257 6.25 and 6.26. Page 324 7.46. Page 394 9.11. Page 398 9.25.
6.15-17 I began with this SAS code:
data patsat; infile '615.dat' firstobs=2; input Satisf Age Severity Anxiety ; proc glm data=patsat; model Satisf = Age Severity Anxiety ; estimate '617a' Intercept 1 Age 35 Severity 45 Anxiety 2.2; output out=anovres r=resid p=fitted; proc print data=anovres;This code produces the anova table, t tests for individual coefficients and the estimates required for predicted values; it also prints out residuals for use later. The output shows:
6.15 c The fitted regression function is
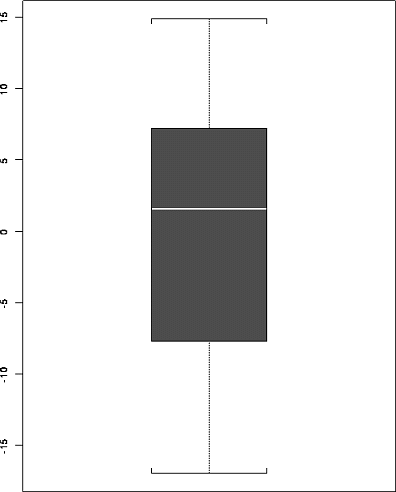
6.15 d Here is a box plot of the (raw) residuals from Splus; I see no problem with outliers.
6.15 e Here is a set of plots from Splus
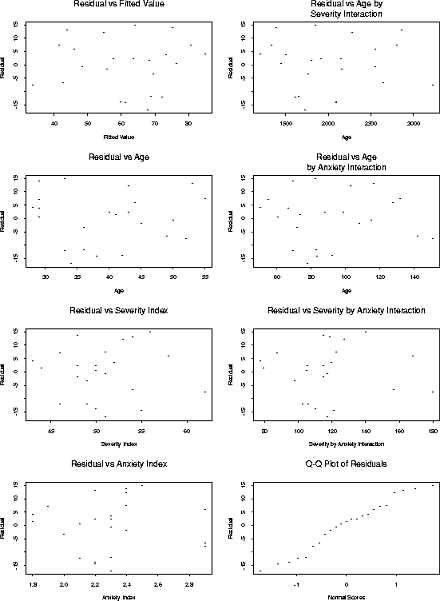
There seems to be no particular problem in any of the plots. The plots show no need for inclusion of the interactions terms and no sign of non-normality.
6.15 f You need replicate observations to compute a pure error sum of squares and you don't have any such. Sometimes people try a clustering technique to split the data set into groups of `near replicates' and then treating these groups as groups of replicates but the technique doesn't work all that well.
6.15 g You have to look in the text for this one. The test regresses
squared residuals on the covariates and computes a ![]() statistic
which looks a lot like an F test (because it was intended to be
analogous to such an F test) except for the numerator not being divided
by degrees of freedom and the denominator being somewhat different; see
page 115 and page 239. I used the code above to print out
a data set which includes the needed residuals. I saved the results
in a file, deleting all the extra output, and then ran this SAS code:
statistic
which looks a lot like an F test (because it was intended to be
analogous to such an F test) except for the numerator not being divided
by degrees of freedom and the denominator being somewhat different; see
page 115 and page 239. I used the code above to print out
a data set which includes the needed residuals. I saved the results
in a file, deleting all the extra output, and then ran this SAS code:
options pagesize=60 linesize=80; data patsatr; infile '615res.dat' firstobs=2; input Obs Satisf Age Severity Anxiety Resid Fitted; rsq=Resid**2; proc glm data=patsatr; model rsq = Age Severity Anxiety ; run;You take the Model Sum of Squares from the output which is 24518 and the Error Sum of Squares from the original output which is 2011.6 and compute
6.16 a The overall F statistic is 13.01 with a P-value of 0.0001 so the
hypothesis that
![]() is rejected at the level 0.1
and, indeed, at any level down to 0.0001. The test implies that at least
one of the three coefficients is not 0.
is rejected at the level 0.1
and, indeed, at any level down to 0.0001. The test implies that at least
one of the three coefficients is not 0.
6.16 b The text intended a joint interval using the Bonferroni procedure: estimate plus or minus t0.05/3,19 times estimated standard errors. The estimates and estimated standard errors are in the SAS output
T for H0: Pr > |T| Std Error of
Parameter Estimate Parameter=0 Estimate
INTERCEPT 162.8758987 6.32 0.0001 25.77565190
AGE -1.2103182 -4.01 0.0007 0.30145159
SEVERITY -0.6659056 -0.81 0.4274 0.82099695
ANXIETY -8.6130315 -0.70 0.4902 12.24125126
The required t critical value is 2.29; you would need to interpolate in
the tables page 1337 between 0.98 and 0.985 since the lower tail area you
actually want is 1-0.05/3=0.98333. Go 2/3 of the way from 2.205 to 2.346.
I actually used Splus.
6.16 c From the output the value of R2 is 0.67. We sometimes describe this as meaning that 2/3 of the variance in patient satisfaction is accounted for by these three covariates. This is a fairly high but not wonderful multiple correlation.
6.17 a The output of the estimate statement is
T for H0: Pr > |T| Std Error of
Parameter Estimate Parameter=0 Estimate
617a 71.6003409 16.11 0.0001 4.44322423
so that the estimate is
9.11 SAS CODE
options pagesize=60 linesize=80;
data patsat;
infile '615.dat' firstobs=2;
input Satisf Age Severity Anxiety ;
proc reg data=patsat;
model Satisf = Age Severity Anxiety /XPX I;
output out=anovres r=resid p=fitted
h=hat dffits=dffits cookd=cookd
rstudent=rstudent press=press;
proc print data=anovres;
The
output
shows that
9.11 a The largest externally studentized residual is for observation 14 at -1.81. This should be compared to the value t0.05/23,19 = 3.2 roughly. (I used the 0.9975 column; you really want 0.9978 so my critical point is a bit too small.) There are no surprising Y outliers.
9.11 b The largest leverage is 0.34 (for observation 9) which should, according to the text (p 377) be compared to 2(4)/23=.35 or so. This is not too large for such a small data set but it would probably warrant a quick look at this point and at case 15 whose leverage is 0.31.
9.11 c You are supposed to compute xT(XTX)-1x when xT = [1,30,58,2]. I printed out the entries in (XTX)-1 (using the I option on the model statement. I used S to compute the desired leverage, getting 0.87 which is an unusually large leverage; I conclude that this would be a substantial extrapolation.
9.11 d The relevant lines of output are
S R
E A S
S V N F T D
A E X I R C P U F
T R I T E O R D F
O I A I E T S O H E E I
B S G T T E I K A S N T
S F E Y Y D D D T S T S
14 51 34 51 2.3 67.9539 -16.9539 0.05661 0.07185 -18.2663 -1.80980 -0.50353
The values of DFFITS and COOKD are not too large; see
Lecture
21
for guidelines. I conclude this data point is ok.
9.11 e I did this partly in S. You need
to compute all the fitted
values with case 14 removed; the easiest way is to delete case 14 from the
data file and rerun. You get the predicted value for case 14 by
subtracting the PRESS residual for case 14 from the true Y for case 14.
I got
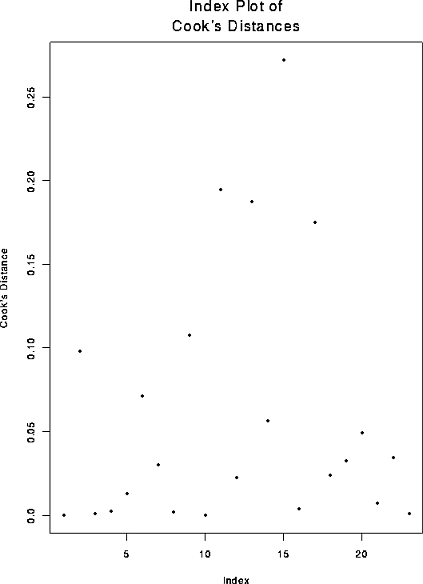
9.11 f You are to plot Di against i. The result is
6.25 You are to choose the ![]() s to minimize
s to minimize
6.26 The multiple correlation coefficient is


7.46 The reduced models are
a)
b)
c)
d)
9.25 If X is invertible then
(XTX)-1 =
X-1[XT]-1 so that
Part B
Solution: We have U=AZ where the matrix A is
![\begin{displaymath}A=\left[\begin{array}{rrr}
\frac{2}{3} & -\frac{1}{3}& - \fra...
...}\\
\frac{1}{3} & \frac{1}{3}& \frac{1}{3}
\end{array}\right]
\end{displaymath}](img30.gif)
![\begin{displaymath}AA^T = \left[\begin{array}{rrr}
\frac{2}{3} & -\frac{1}{3}& 0...
...}& \frac{2}{3} & 0 \\
0 & 0 & \frac{1}{3}
\end{array}\right]
\end{displaymath}](img31.gif)
Solution: The variance covariance matrix in the previous part is block diagonal with a 2 by 2 block and a 1 by 1 block, so the first two components are independent of the last component.
Solution:
![]() .
Now
.
Now
![]() so
so
Solution: The function G is
Solution: This question is wrong! You can write
Solution: The density in class was
![\begin{displaymath}\frac{1}{2\pi\sqrt{det(AA^T)}}\exp\left[-\frac{1}{2} (x -\mu)^T
\Sigma^{-1} (x-\mu)\right]
\end{displaymath}](img43.gif)
The entries in ![]() are c and f and we have
are c and f and we have
![\begin{displaymath}A = \left[\begin{array}{rr} a & b \\ d & e \end{array}\right]
\end{displaymath}](img46.gif)
![\begin{displaymath}\Sigma = AA^T = \left[\begin{array}{cc} a^2+b^2 & ad+be \\ ad+be & d^2+e^2
\end{array}\right]
\end{displaymath}](img47.gif)
![\begin{displaymath}(AA^T)^{-1} = \left[\begin{array}{cc}
\frac{d^2+e^2}{(ae-bd)^...
...+be}{(ae-bd)^2} & \frac{a^2+b^2}{(ae-bd)^2}
\end{array}\right]
\end{displaymath}](img48.gif)
![\begin{displaymath}f(x_1,x_2) = \frac{1}{2\pi\vert ae-bd\vert}
\exp\left[-\frac{1}{2} \frac{q(x_1,x_2,a,b,c,d,e,f)}{(ae-bd)^2}\right] \, .
\end{displaymath}](img49.gif)
![\begin{eqnarray*}q(x_1,x_2,a,b,c,d,e,f) & = &
[(x_1-c)^2 (d^2+e^2) \\
& & -2(ad+be)(x_1-c)(x_2-f) \\
& & + (x_2-f)^2 ( a^2+b^2)] \,.
\end{eqnarray*}](img50.gif)
To compute
![]() we take the joint density of X1 and X2and integrate it over the set of (x1,x2) such that
we take the joint density of X1 and X2and integrate it over the set of (x1,x2) such that ![]() to get
to get