 . Idea: study structure of
. Idea: study structure of
Suppose
 . Idea: study structure of
. Idea: study structure of
![]() or of
or of ![]() , the correlation matrix derived from
, the correlation matrix derived from
![]() .
.
Which linear combination of the entries of ![]() has
maximal variance?
has
maximal variance?
None: variance of
![]() is
is
![]() which is proportional to
squared length of
which is proportional to
squared length of ![]() .
.
So: maximize

Now: find another linear combination which is independent of
first and maximizes variance (divided by
![]() ).
).
Principal Components: Find
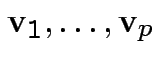 which are orthonormal eigenvectors of
which are orthonormal eigenvectors of
![]() . Let
. Let ![]() ,
,
 be the corresponding eigenvalues. We call
be the corresponding eigenvalues. We call
 the principal components of
the principal components of
![]() .
.
If ![]() has columns
has columns ![]() then
then
![]() has a MVN
has a MVN
 where
where
![]() is diagonal
matrix of eigenvalues.
is diagonal
matrix of eigenvalues.
Columns of
![]() are the components.
are the components.
We can reconstruct the original ![]() from the components
from the components
 because
because


Points to ponder:
1: Principal component ![]() has variance
has variance ![]() .
.
2: Can compare ![]() to
to
 .
.
3: Try to interpret components; often leading component is some sort of average then others are contrasts.
4: To estimate replace
![]() by
by ![]() .
.
5: Process can be done on original ![]() or on standardized
variates:
or on standardized
variates:

6: General advice: use ![]() when different variables in
when different variables in
![]() are commensurate (same units, comparable). Use
are commensurate (same units, comparable). Use ![]() otherwise.
otherwise.
Data: Table 9.12 in Johnson and Wichern.
3 measurements on sales performance and 4 test scorese for 50 salespeople in large firm. The data begin
| Growth | Profit | New | Creat | Mech | Abst | Math |
| 93.0 | 96.0 | 97.8 | 9 | 12 | 9 | 20 |
| 88.8 | 91.8 | 96.8 | 7 | 10 | 10 | 15 |
| 95.0 | 100.3 | 99.0 | 8 | 12 | 9 | 26 |
Read in and print out the data.
> sales <- matrix(scan("T9-12.DAT"),ncol=7,byrow=T)
> sales
[,1] [,2] [,3] [,4] [,5] [,6] [,7]
[1,] 93.0 96.0 97.8 9 12 9 20
[2,] 88.8 91.8 96.8 7 10 10 15
[3,] 95.0 100.3 99.0 8 12 9 26
Compute the variance covariance matrix and the correlation matrix and
print them out.
> S <- var(sales) > S 53.837 68.794 30.564 16.580 17.585 10.588 71.699 68.794 102.502 40.195 21.656 25.561 10.081 100.744 30.565 40.195 22.205 13.037 10.168 6.464 42.335 16.580 21.656 13.037 15.604 7.898 1.242 17.176 17.585 25.561 10.168 7.898 11.457 2.795 20.493 10.588 10.081 6.464 1.242 2.795 4.578 12.770 71.698 100.744 42.335 17.176 20.493 12.770 111.043 > D <- diag(sqrt(diag(S))) > D 7.33735 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.00000 10.1243 0.0000 0.0000 0.0000 0.0000 0.0000 0.00000 0.0000 4.7122 0.0000 0.0000 0.0000 0.0000 0.00000 0.0000 0.0000 3.9502 0.0000 0.0000 0.0000 0.00000 0.0000 0.0000 0.0000 3.3848 0.0000 0.0000 0.00000 0.0000 0.0000 0.0000 0.0000 2.1396 0.0000 0.00000 0.0000 0.0000 0.0000 0.0000 0.0000 10.5377 > R <- solve(D,S)%*% solve(D)
> R
1.0000 0.9261 0.8840 0.5720 0.7081 0.6741 0.9273
0.9261 1.0000 0.8425 0.5415 0.7459 0.4654 0.9443
0.8840 0.8425 1.0000 0.7004 0.6375 0.6411 0.8526
0.5720 0.5415 0.7004 1.0000 0.5907 0.1469 0.4126
0.7081 0.7459 0.6375 0.5907 1.0000 0.3860 0.5746
0.6744 0.4654 0.6411 0.1469 0.3860 1.0000 0.5664
0.9273 0.9443 0.8526 0.4126 0.5746 0.5664 1.0000
> round(R,2)
[,1] [,2] [,3] [,4] [,5] [,6] [,7]
[1,] 1.00 0.93 0.88 0.57 0.71 0.67 0.93
[2,] 0.93 1.00 0.84 0.54 0.75 0.47 0.94
[3,] 0.88 0.84 1.00 0.70 0.64 0.64 0.85
[4,] 0.57 0.54 0.70 1.00 0.59 0.15 0.41
[5,] 0.71 0.75 0.64 0.59 1.00 0.39 0.57
[6,] 0.67 0.47 0.64 0.15 0.39 1.00 0.57
[7,] 0.93 0.94 0.85 0.41 0.57 0.57 1.00
Correlations between three measures of sales performance -
all quite high.
Also very high: correlation of math score with the three sales performance indices.
I find it interesting that the correlations amongst the 4 test scores are not all that high.
Compute eigenvalues and eigenvectors of ![]() .
.
Note: without symmetric=T argument to eigen might not get normalized eigenvectors.
> e.S <- eigen(S,symmetric=T)
> e.S
$values:
[1] 285.1366314 17.2678388 8.9785502 5.8957342
2.5247046 1.1505297
[7] 0.2710316
$vectors:
[,1] [,2] [,3] [,4] [,5] [,6]
0.42055 -0.12038 -0.31712 -0.47136 0.59884 0.05743
0.58892 -0.06545 0.59396 0.07524 -0.05950 0.53029
0.25070 -0.25607 -0.46093 0.16111 -0.62886 0.17742
0.12864 -0.73755 -0.15906 0.41980 0.25206 -0.14283
0.14144 -0.36484 0.36098 -0.52253 -0.34536 -0.56932
0.07253 0.05320 -0.38705 -0.47051 -0.24056 0.30162
0.60962 0.48553 -0.16961 0.27486 -0.04146 -0.49852
[,7]
0.35212680
-0.07541783
0.45411767
-0.39244052
0.01215659
-0.68693393
-0.19509084
Store the eigenvectors in ![]() and check that
and check that ![]() .
.
> P.S <- e.S$vectors
> P.S %*% t(P.S)
[,1] [,2] [,3] [,4] [,5]
1.000e+00 3.990e-16 8.327e-17 3.886e-16 -5.117e-17
3.990e-16 1.000e+00 -6.939e-18 1.492e-16 1.315e-16
8.327e-17 -6.939e-18 1.000e+00 3.886e-16 -1.830e-16
3.886e-16 1.492e-16 3.886e-16 1.000e+00 3.452e-16
-5.117e-17 1.315e-16 -1.830e-16 3.452e-16 1.000e+00
-9.159e-16 3.053e-16 0.000e+00 -5.551e-17 -3.036e-16
1.804e-16 -1.006e-16 2.776e-17 -3.608e-16 -1.518e-17
[,6] [,7]
[1,] -9.159340e-16 1.804112e-16
[2,] 3.053113e-16 -1.006140e-16
[3,] 0.000000e+00 2.775558e-17
[4,] -5.551115e-17 -3.608225e-16
[5,] -3.035766e-16 -1.517883e-17
[6,] 1.000000e+00 -5.551115e-17
[7,] -5.551115e-17 1.000000e+00
> round(.Last.value,6)
[,1] [,2] [,3] [,4] [,5] [,6] [,7]
[1,] 1 0 0 0 0 0 0
[2,] 0 1 0 0 0 0 0
[3,] 0 0 1 0 0 0 0
[4,] 0 0 0 1 0 0 0
[5,] 0 0 0 0 1 0 0
[6,] 0 0 0 0 0 1 0
[7,] 0 0 0 0 0 0 1
Put eigenvalues in diagonal matrix > L.S <- diag(e.S$values) > L.S [,1] [,2] [,3] [,4] [,5] [,6] [,7] 285.137 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.000 17.2678 0.0000 0.0000 0.0000 0.0000 0.0000 0.000 0.0000 8.9786 0.0000 0.0000 0.0000 0.0000 0.000 0.0000 0.0000 5.8957 0.0000 0.0000 0.0000 0.000 0.0000 0.0000 0.0000 2.5247 0.0000 0.0000 0.000 0.0000 0.0000 0.0000 0.0000 1.1505 0.0000 0.000 0.0000 0.0000 0.0000 0.0000 0.0000 0.2710Check the identity
> P.S%*% L.S %*% t(P.S) - S
[,1] [,2] [,3] [,4] [,5]
-2.132e-14 -2.842e-14 -1.066e-14 1.421e-14 -3.553e-15
-2.842e-14 -9.948e-14 -3.553e-14 3.553e-15 -1.421e-14
-7.105e-15 -2.842e-14 -7.105e-15 1.243e-14 -5.329e-15
1.776e-14 7.105e-15 1.243e-14 1.243e-14 8.882e-15
-3.553e-15 -1.421e-14 -3.553e-15 8.882e-15 -7.105e-15
-8.527e-14 -7.816e-14 -4.263e-14 -9.992e-15 -2.753e-14
2.842e-14 -2.842e-14 0.000e+00 3.553e-15 -3.553e-15
[,6] [,7]
-8.348877e-14 1.421085e-14
-7.815970e-14 -4.263256e-14
-4.263256e-14 0.000000e+00
-9.992007e-15 3.552714e-15
-2.664535e-14 0.000000e+00
-5.240253e-14 -3.907985e-14
-3.730349e-14 5.684342e-14
Watch what happens if you don't use symmetric=T.
> e.R <- eigen(R)
> e.R
$values:
[1] 5.03459779 0.93351614 0.49791975
0.42124549 0.08104043 0.02034063
0.01133977
$vectors:
et cetera
> P.R <- e.R$vectors
> P.R %*% t(P.R)
[,1] [,2] [,3] [,4] [,5] [,6] [,7]
1.3135 0.3704 0.1903 0.3166 0.2481 0.3133 0.3503
0.3704 1.0729 0.3820 0.2968 0.3079 0.2920 0.3169
0.1903 0.3820 1.3334 0.1737 0.3199 0.1931 0.3926
0.3166 0.2968 0.1737 1.0804 0.2238 -0.0134 0.2442
0.2481 0.3079 0.3199 0.2238 1.1357 0.1553 0.2502
0.3133 0.2920 0.1931 -0.0134 0.1553 0.9978 0.3284
0.3503 0.3169 0.3926 0.2442 0.2502 0.3284 1.0804
This should have been the identity but isn't.
Now make a plot of the first principal component versus the second.
> Y.S <- sales %*% P.S
> postscript("prcmps_S.ps",horizontal=F)
> plot(Y.S[,1],Y.S[,2],xlab="First PC",
ylab="Second PC",main="Plot of 1st
vs 2nd PC\n us ing S")
> dev.off()
Generated postscript file "prcmps_S.ps".

> P.S
[,1] [,2] [,3] [,4] [,5] [,6]
0.4206 -0.1204 -0.3171 -0.4714 0.5988 0.0574
0.5889 -0.0654 0.5940 0.0752 -0.0595 0.5309
0.2507 -0.2561 -0.4609 0.1611 -0.6289 0.1774
0.1286 -0.7375 -0.1591 0.4198 0.2521 -0.1428
0.1414 -0.3648 0.3610 -0.5225 -0.3454 -0.5693
0.0725 0.0532 -0.3871 -0.4705 -0.2406 0.3016
0.6096 0.4855 -0.1696 0.2749 -0.0415 -0.4985
[,7]
0.35212680
-0.07541783
0.45411767
-0.39244052
0.01215659
-0.68693393
-0.19509084
Leading principal component: used first column of Hard to interpret although all the weights are positive.
Big weights on components with large variances which is natural but uninformative.
The trouble is that the 7 variables are not all really comparable.
Do whole thing again with
![]() .
.
> e.R <- eigen(R,symmetric=T)
> e.R
$values:
[1] 5.03459779 0.93351614 0.49791975
0.42124549 0.08104043 0.02034063
0.01133977
$vectors:
[,1] [,2] [,3] [,4] [,5] [,6]
0.4337 0.1118 -0.0755 0.0424 -0.6325 -0.3366
0.4202 -0.0293 -0.4425 -0.0108 0.0001 0.7853
0.4211 -0.0092 0.2042 0.3249 0.7010 -0.1568
0.2943 -0.6684 0.4515 0.3027 -0.2610 0.1142
0.3491 -0.2950 0.0059 -0.8466 0.1742 -0.1970
0.2892 0.6424 0.6038 -0.1537 -0.0870 0.2363
0.4074 0.2004 -0.4340 0.2460 0.0496 -0.3711
[,7]
[1,] 0.52782527
[2,] 0.09948330
[3,] 0.39916419
[4,] -0.29995962
[5,] -0.07231139
[6,] -0.22844351
[7,] -0.63622351
> P.R <- e.R$vectors
> L.R <- diag(e.R$values)
> L.R
[,1] [,2] [,3] [,4] [,5] [,6] [,7]
[1,] 5.035 0.000 0.000 0.000 0.000 0.000 0.000
[2,] 0.000 0.934 0.000 0.000 0.000 0.000 0.000
[3,] 0.000 0.000 0.498 0.000 0.000 0.000 0.000
[4,] 0.000 0.000 0.000 0.421 0.000 0.000 0.000
[5,] 0.000 0.000 0.000 0.000 0.081 0.000 0.000
[6,] 0.000 0.000 0.000 0.000 0.000 0.020 0.000
[7,] 0.000 0.000 0.000 0.000 0.000 0.000 0.011
Try to interpret eigenvectors of
> P.R
[,1] [,2] [,3] [,4] [,5] [,6]
0.433 0.112 -0.075 0.042 -0.632 -0.337
0.420 -0.029 -0.442 -0.011 0.000 0.785
0.421 -0.009 0.204 0.325 0.701 -0.157
0.294 -0.668 0.451 0.303 -0.261 0.114
0.349 -0.295 0.006 -0.847 0.174 -0.197
0.289 0.642 0.604 -0.154 -0.087 0.236
0.407 0.200 -0.434 0.246 0.050 -0.371
[,7]
[1,] 0.52782527
[2,] 0.09948330
[3,] 0.39916419
[4,] -0.29995962
[5,] -0.07231139
[6,] -0.22844351
[7,] -0.63622351
Entries in first column nearly constant: leading
principal component is close to just being average of 7 variables,
each expressed in standard units (
 ).
).
Next principal component is essentially a weighted average of Mathematics and Abstract Reasoning minus a similar average of Mechanical reasoning and Creativity.
The third PC seems hard to interpret.
Now transform the data to get the principal components
 .
Then plot the first two principal components against each other and
examine the plot looking for interesting cases.
.
Then plot the first two principal components against each other and
examine the plot looking for interesting cases.
> postscript("prcmps_R.ps", horizontal = F)
> plot(Y.R[, 1], Y.R[, 2], xlab = "First PC",
ylab = "Second PC", main =
"Plot of 1st vs 2nd PC\n using R")
> l1 <- (abs(Y.R[,1])>25)
> l1
[1] F F F F F F F T F F F F F F F T F F F
F F F F F F F F F F F F F F F F F F F
F F F F F T F F F T F F
> l2 <- (abs(Y.R[,2]) > 6)
> l2
[1] F F F F F F F F F F F F F F F F F F F
F F T F F F F F F F F F T F F F T T F
F F F T F F F F F F F F
> l3 <- (l1|l2)
> l3
[1] F F F F F F F T F F F F F F F T F F F
F F T F F F F F F F F F T F F F T T F
F F F T F T F F F T F F
> text(Y.R[,1],Y.R[,2],labels=l3)
> dev.off()
Generated postscript file "prcmps_R.ps".
null device
1
Here is the plot.