Principal Components example
The data for this example are in Table 9.12 in Johnson and Wichern. They consist of 3 measurements on the sales performance of 50 salespeople for a large firm and 4 test scores. The data begin
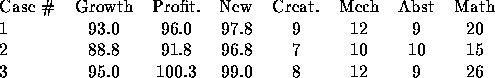
In what follows I do PCA on both the sample variance covariance matrix and on the sample correlation matrix. The S code is included as a tutorial for those interested. Read in and print out the data.
> sales <- matrix(scan("T9-12.DAT"),ncol=7,byrow=T)
> sales
[,1] [,2] [,3] [,4] [,5] [,6] [,7]
[1,] 93.0 96.0 97.8 9 12 9 20
[2,] 88.8 91.8 96.8 7 10 10 15
[3,] 95.0 100.3 99.0 8 12 9 26
Compute the variance covariance matrix and the correlation matrix and
print them out.
> S <- var(sales)
> S
[,1] [,2] [,3] [,4] [,5] [,6] [,7]
[1,] 53.83664 68.79409 30.564531 16.579673 17.585224 10.587592 71.69861
[2,] 68.79409 102.50175 40.195082 21.656286 25.561265 10.081306 100.74416
[3,] 30.56453 40.19508 22.205000 13.036531 10.167551 6.463673 42.33510
[4,] 16.57967 21.65629 13.036531 15.603673 7.898367 1.241633 17.17633
[5,] 17.58522 25.56127 10.167551 7.898367 11.456735 2.795102 20.49306
[6,] 10.58759 10.08131 6.463673 1.241633 2.795102 4.577959 12.76980
[7,] 71.69861 100.74416 42.335102 17.176327 20.493061 12.769796 111.04327
> D <- diag(sqrt(diag(S)))
> D
[,1] [,2] [,3] [,4] [,5] [,6] [,7]
[1,] 7.337345 0.00000 0.000000 0.000000 0.00000 0.000000 0.00000
[2,] 0.000000 10.12431 0.000000 0.000000 0.00000 0.000000 0.00000
[3,] 0.000000 0.00000 4.712218 0.000000 0.00000 0.000000 0.00000
[4,] 0.000000 0.00000 0.000000 3.950149 0.00000 0.000000 0.00000
[5,] 0.000000 0.00000 0.000000 0.000000 3.38478 0.000000 0.00000
[6,] 0.000000 0.00000 0.000000 0.000000 0.00000 2.139617 0.00000
[7,] 0.000000 0.00000 0.000000 0.000000 0.00000 0.000000 10.53771
> R <- solve(D,S)%*% solve(D)
> R
[,1] [,2] [,3] [,4] [,5] [,6] [,7]
[1,] 1.0000000 0.9260758 0.8840023 0.5720363 0.7080738 0.6744073 0.9273116
[2,] 0.9260758 1.0000000 0.8425232 0.5415080 0.7459097 0.4653880 0.9442960
[3,] 0.8840023 0.8425232 1.0000000 0.7003630 0.6374712 0.6410886 0.8525682
[4,] 0.5720363 0.5415080 0.7003630 1.0000000 0.5907360 0.1469074 0.4126395
[5,] 0.7080738 0.7459097 0.6374712 0.5907360 1.0000000 0.3859502 0.5745533
[6,] 0.6744073 0.4653880 0.6410886 0.1469074 0.3859502 1.0000000 0.5663721
[7,] 0.9273116 0.9442960 0.8525682 0.4126395 0.5745533 0.5663721 1.0000000
> round(R,2)
[,1] [,2] [,3] [,4] [,5] [,6] [,7]
[1,] 1.00 0.93 0.88 0.57 0.71 0.67 0.93
[2,] 0.93 1.00 0.84 0.54 0.75 0.47 0.94
[3,] 0.88 0.84 1.00 0.70 0.64 0.64 0.85
[4,] 0.57 0.54 0.70 1.00 0.59 0.15 0.41
[5,] 0.71 0.75 0.64 0.59 1.00 0.39 0.57
[6,] 0.67 0.47 0.64 0.15 0.39 1.00 0.57
[7,] 0.93 0.94 0.85 0.41 0.57 0.57 1.00
Look at the correlations between the three measures of sales performance -
all are quite high. Also very high is the correlation of math score with
the three sales performance indices. I find it interesting that the
correlations amongst the 4 test scores are not all that high.
Compute the eigenvalues and eigenvectors of S. Note that without the symmetric=T argument to eigen you might not get normalized eigenvectors.
> e.S <- eigen(S,symmetric=T)
> e.S
$values:
[1] 285.1366314 17.2678388 8.9785502 5.8957342 2.5247046 1.1505297
[7] 0.2710316
$vectors:
[,1] [,2] [,3] [,4] [,5] [,6]
[1,] 0.42055360 -0.12038402 -0.3171150 -0.47136100 0.59883830 0.05743392
[2,] 0.58891939 -0.06544701 0.5939649 0.07524351 -0.05949667 0.53028957
[3,] 0.25070446 -0.25606621 -0.4609316 0.16111410 -0.62885652 0.17742124
[4,] 0.12864131 -0.73754639 -0.1590590 0.41980231 0.25205680 -0.14283133
[5,] 0.14144240 -0.36483937 0.3609846 -0.52252896 -0.34535826 -0.56931521
[6,] 0.07253396 0.05320196 -0.3870509 -0.47051219 -0.24055838 0.30161519
[7,] 0.60962298 0.48553226 -0.1696108 0.27485861 -0.04146119 -0.49852152
[,7]
[1,] 0.35212680
[2,] -0.07541783
[3,] 0.45411767
[4,] -0.39244052
[5,] 0.01215659
[6,] -0.68693393
[7,] -0.19509084
Store the eigenvectors in P and check that > P.S <- e.S$vectors
> P.S %*% t(P.S)
[,1] [,2] [,3] [,4] [,5]
[1,] 1.000000e+00 3.989864e-16 8.326673e-17 3.885781e-16 -5.117434e-17
[2,] 3.989864e-16 1.000000e+00 -6.938894e-18 1.491862e-16 1.315137e-16
[3,] 8.326673e-17 -6.938894e-18 1.000000e+00 3.885781e-16 -1.830133e-16
[4,] 3.885781e-16 1.491862e-16 3.885781e-16 1.000000e+00 3.452100e-16
[5,] -5.117434e-17 1.315137e-16 -1.830133e-16 3.452100e-16 1.000000e+00
[6,] -9.159340e-16 3.053113e-16 0.000000e+00 -5.551115e-17 -3.035766e-16
[7,] 1.804112e-16 -1.006140e-16 2.775558e-17 -3.608225e-16 -1.517883e-17
[,6] [,7]
[1,] -9.159340e-16 1.804112e-16
[2,] 3.053113e-16 -1.006140e-16
[3,] 0.000000e+00 2.775558e-17
[4,] -5.551115e-17 -3.608225e-16
[5,] -3.035766e-16 -1.517883e-17
[6,] 1.000000e+00 -5.551115e-17
[7,] -5.551115e-17 1.000000e+00
> round(.Last.value,6)
[,1] [,2] [,3] [,4] [,5] [,6] [,7]
[1,] 1 0 0 0 0 0 0
[2,] 0 1 0 0 0 0 0
[3,] 0 0 1 0 0 0 0
[4,] 0 0 0 1 0 0 0
[5,] 0 0 0 0 1 0 0
[6,] 0 0 0 0 0 1 0
[7,] 0 0 0 0 0 0 1
Put the eigenvalues in a diagonal matrix > L.S <- diag(e.S$values)
> L.S
[,1] [,2] [,3] [,4] [,5] [,6] [,7]
[1,] 285.1366 0.00000 0.00000 0.000000 0.000000 0.00000 0.0000000
[2,] 0.0000 17.26784 0.00000 0.000000 0.000000 0.00000 0.0000000
[3,] 0.0000 0.00000 8.97855 0.000000 0.000000 0.00000 0.0000000
[4,] 0.0000 0.00000 0.00000 5.895734 0.000000 0.00000 0.0000000
[5,] 0.0000 0.00000 0.00000 0.000000 2.524705 0.00000 0.0000000
[6,] 0.0000 0.00000 0.00000 0.000000 0.000000 1.15053 0.0000000
[7,] 0.0000 0.00000 0.00000 0.000000 0.000000 0.00000 0.2710316
Check the identity > P.S%*% L.S %*% t(P.S) - S
[,1] [,2] [,3] [,4] [,5]
[1,] -2.131628e-14 -2.842171e-14 -1.065814e-14 1.421085e-14 -3.552714e-15
[2,] -2.842171e-14 -9.947598e-14 -3.552714e-14 3.552714e-15 -1.421085e-14
[3,] -7.105427e-15 -2.842171e-14 -7.105427e-15 1.243450e-14 -5.329071e-15
[4,] 1.776357e-14 7.105427e-15 1.243450e-14 1.243450e-14 8.881784e-15
[5,] -3.552714e-15 -1.421085e-14 -3.552714e-15 8.881784e-15 -7.105427e-15
[6,] -8.526513e-14 -7.815970e-14 -4.263256e-14 -9.992007e-15 -2.753353e-14
[7,] 2.842171e-14 -2.842171e-14 0.000000e+00 3.552714e-15 -3.552714e-15
[,6] [,7]
[1,] -8.348877e-14 1.421085e-14
[2,] -7.815970e-14 -4.263256e-14
[3,] -4.263256e-14 0.000000e+00
[4,] -9.992007e-15 3.552714e-15
[5,] -2.664535e-14 0.000000e+00
[6,] -5.240253e-14 -3.907985e-14
[7,] -3.730349e-14 5.684342e-14
Watch what happens if you don't use symmetric=T.
> e.R <- eigen(R)
> e.R
$values:
[1] 5.03459779 0.93351614 0.49791975 0.42124549 0.08104043 0.02034063
0.01133977
$vectors:
[,1] [,2] [,3] [,4] [,5]
[,6]
[1,] 0.7269274 -0.116207095 -0.056760689 0.03974777 -0.687192227
-0.28993184
[2,] 0.7043683 0.030454407 -0.332704938 -0.01008627 0.000128432
0.67646542
[3,] 0.7057720 0.009568613 0.153532259 0.30479417 0.761650850
-0.13507161
[4,] 0.4932870 0.695047750 0.339482201 0.28395450 -0.283579937
0.09834274
[5,] 0.5851533 0.306695959 0.004452648 -0.79414371 0.189333749
-0.16961040
[6,] 0.4847059 -0.667972463 0.453988739 -0.14415168 -0.094478767
0.20350664
[7,] 0.6828969 -0.208350975 -0.326359275 0.23076897 0.053870538
-0.31966116
[,7]
[1,] -0.45877388
[2,] -0.08646865
[3,] -0.34694455
[4,] 0.26071817
[5,] 0.06285144
[6,] 0.19855797
[7,] 0.55299119
> P.R <- e.R$vectors
> P.R %*% t(P.R)
[,1] [,2] [,3] [,4] [,5] [,6]
[,7]
[1,] 1.3134963 0.3704216 0.1902634 0.3165822 0.2481377 0.3132995
0.3502879
[2,] 0.3704216 1.0729387 0.3819860 0.2967566 0.3078862 0.2919624
0.3168702
[3,] 0.1902634 0.3819860 1.3334041 0.1737410 0.3198625 0.1931292
0.3925566
[4,] 0.3165822 0.2967566 0.1737410 1.0803646 0.2238431 -0.0134117
0.2442469
[5,] 0.2481377 0.3078862 0.3198625 0.2238431 1.1357161 0.1553362
0.2501557
[6,] 0.3132995 0.2919624 0.1931292 -0.0134117 0.1553362 0.9977790
0.3284057
[7,] 0.3502879 0.3168702 0.3925566 0.2442469 0.2501557 0.3284057
1.0804075
This should have been the identity but isn't. Now make a plot of the
first principal component versus the second.
> Y.S <- sales %*% P.S
> postscript("prcmps_S.ps",horizontal=F)
> plot(Y.S[,1],Y.S[,2],xlab="First PC",ylab="Second PC",main="Plot of 1st
vs 2nd PC\n us ing S")
> dev.off()
Generated postscript file "prcmps_S.ps".
Now try to interpret the eigenvectors
> P.S
[,1] [,2] [,3] [,4] [,5] [,6]
[1,] 0.42055360 -0.12038402 -0.3171150 -0.47136100 0.59883830 0.05743392
[2,] 0.58891939 -0.06544701 0.5939649 0.07524351 -0.05949667 0.53028957
[3,] 0.25070446 -0.25606621 -0.4609316 0.16111410 -0.62885652 0.17742124
[4,] 0.12864131 -0.73754639 -0.1590590 0.41980231 0.25205680 -0.14283133
[5,] 0.14144240 -0.36483937 0.3609846 -0.52252896 -0.34535826 -0.56931521
[6,] 0.07253396 0.05320196 -0.3870509 -0.47051219 -0.24055838 0.30161519
[7,] 0.60962298 0.48553226 -0.1696108 0.27485861 -0.04146119 -0.49852152
[,7]
[1,] 0.35212680
[2,] -0.07541783
[3,] 0.45411767
[4,] -0.39244052
[5,] 0.01215659
[6,] -0.68693393
[7,] -0.19509084
The leading principal component used the first column of P as
coefficients. It is hard to interpret although all the weights are
positive. The big weights are on the components with large variances
which is natural but uninformative. The trouble is that the 7 variables
are not all really comparable. Now we do the whole thing again with
R.
> e.R <- eigen(R,symmetric=T)
> e.R
$values:
[1] 5.03459779 0.93351614 0.49791975 0.42124549 0.08104043 0.02034063
0.01133977
$vectors:
[,1] [,2] [,3] [,4] [,5] [,6]
[1,] 0.4336719 0.111754422 -0.075488541 0.04237344 -0.6324942624 -0.3365963
[2,] 0.4202136 -0.029287495 -0.442478953 -0.01075255 0.0001182093 0.7853424
[3,] 0.4210510 -0.009201975 0.204189315 0.32492838 0.7010262539 -0.1568114
[4,] 0.2942863 -0.668415809 0.451492333 0.30271208 -0.2610080204 0.1141710
[5,] 0.3490920 -0.294944379 0.005921773 -0.84660356 0.1742634819 -0.1969091
[6,] 0.2891669 0.642377957 0.603779622 -0.15367411 -0.0869586057 0.2362610
[7,] 0.4074041 0.200367651 -0.434039576 0.24601320 0.0495826418 -0.3711105
[,7]
[1,] 0.52782527
[2,] 0.09948330
[3,] 0.39916419
[4,] -0.29995962
[5,] -0.07231139
[6,] -0.22844351
[7,] -0.63622351
> P.R <- e.R$vectors
> L.R <- diag(e.R$values)
> L.R
[,1] [,2] [,3] [,4] [,5] [,6] [,7]
[1,] 5.034598 0.0000000 0.0000000 0.0000000 0.00000000 0.00000000 0.00000000
[2,] 0.000000 0.9335161 0.0000000 0.0000000 0.00000000 0.00000000 0.00000000
[3,] 0.000000 0.0000000 0.4979197 0.0000000 0.00000000 0.00000000 0.00000000
[4,] 0.000000 0.0000000 0.0000000 0.4212455 0.00000000 0.00000000 0.00000000
[5,] 0.000000 0.0000000 0.0000000 0.0000000 0.08104043 0.00000000 0.00000000
[6,] 0.000000 0.0000000 0.0000000 0.0000000 0.00000000 0.02034063 0.00000000
[7,] 0.000000 0.0000000 0.0000000 0.0000000 0.00000000 0.00000000 0.01133977
Now lets try to interpret the eigenvectors for the correlation matrix.
> P.R
[,1] [,2] [,3] [,4] [,5] [,6]
[1,] 0.4336719 0.111754422 -0.075488541 0.04237344 -0.6324942624 -0.3365963
[2,] 0.4202136 -0.029287495 -0.442478953 -0.01075255 0.0001182093 0.7853424
[3,] 0.4210510 -0.009201975 0.204189315 0.32492838 0.7010262539 -0.1568114
[4,] 0.2942863 -0.668415809 0.451492333 0.30271208 -0.2610080204 0.1141710
[5,] 0.3490920 -0.294944379 0.005921773 -0.84660356 0.1742634819 -0.1969091
[6,] 0.2891669 0.642377957 0.603779622 -0.15367411 -0.0869586057 0.2362610
[7,] 0.4074041 0.200367651 -0.434039576 0.24601320 0.0495826418 -0.3711105
[,7]
[1,] 0.52782527
[2,] 0.09948330
[3,] 0.39916419
[4,] -0.29995962
[5,] -0.07231139
[6,] -0.22844351
[7,] -0.63622351
The entries in the first column are nearly constant so that the leading
principal component is close to just being an average of the 7 variables,
each expressed in standard units (
Now transform the data to get the principal components ![]() .
Then plot the first two principal components against each other and
examine the plot looking for interesting cases.
.
Then plot the first two principal components against each other and
examine the plot looking for interesting cases.
> postscript("prcmps_R.ps", horizontal = F)
> plot(Y.R[, 1], Y.R[, 2], xlab = "First PC", ylab = "Second PC", main =
+ "Plot of 1st vs 2nd PC\n using R")
> l1 <- (abs(Y.R[,1])>25)
> l1
[1] F F F F F F F T F F F F F F F T F F F F F F F F F F F F F F F F F F F
F F F
[39] F F F F F T F F F T F F
> l2 <- (abs(Y.R[,2]) > 6)
> l2
[1] F F F F F F F F F F F F F F F F F F F F F T F F F F F F F F F T F F F
T T F
[39] F F F T F F F F F F F F
> l3 <- (l1|l2)
> l3
[1] F F F F F F F T F F F F F F F T F F F F F T F F F F F F F F F T F F F
T T F
[39] F F F T F T F F F T F F
> text(Y.R[,1],Y.R[,2],labels=l3)
> dev.off()
Generated postscript file "prcmps_R.ps".
null device
1
Here is the plot.
