PROC FASTCLUS Statement
- PROC FASTCLUS MAXCLUSTERS= n |
RADIUS=t < options > ;
You must specify either the MAXCLUSTERS= or the RADIUS= argument in
the PROC FASTCLUS statement.
- MAXCLUSTERS=n
- MAXC=n
-
specifies the maximum number of clusters allowed.
If you omit the MAXCLUSTERS= option, a value of 100 is assumed.
- RADIUS=t
- R=t
-
establishes the minimum distance criterion for selecting new seeds.
No observation is considered as a new seed
unless its minimum distance to previous seeds
exceeds the value given by the RADIUS= option.
The default value is 0.
If you specify the REPLACE=RANDOM option, the RADIUS= option is ignored.
You can specify the following options
in the PROC FASTCLUS statement.
Table 27.1 summarizes the
options.
Table 27.1: Options Available in the PROC FASTCLUS Statement
|
Task
|
|
Options
|
| Specify data set details | | CLUSTER= |
| | | DATA= |
| | | MEAN= |
| | | OUT= |
| | | OUTITER |
| | | OUTSEED= |
| | | OUTSTAT= |
| | | SEED= |
| Specify distance dimension | | BINS= |
| | | HC= |
| | | HP= |
| | | IRLS |
| | | LEAST= |
| Select initial cluster seeds | | RANDOM= |
| | | REPLACE= |
| Compute final cluster seeds | | CONVERGE= |
| | | DELETE= |
| | | DRIFT |
| | | MAXCLUSTERS= |
| | | MAXITER= |
| | | RADIUS= |
| | | STRICT |
| Work with missing values | | IMPUTE |
| | | NOMISS |
| Specify variance divisor | | VARDEF |
| Control output | | DISTANCE |
| | | LIST |
| | | NOPRINT |
| | | SHORT |
| | | SUMMARY |
The following list provides details on these options.
The list is in alphabetical order.
- BINS=n
-
specifies the number of bins used in the
bin-sort algorithm for computing medians for LEAST=1.
By default, PROC FASTCLUS uses from 10 to 100 bins,
depending on the amount of memory available.
Larger values use more memory and make each iteration
somewhat slower, but they may reduce the number of iterations.
Smaller values have the opposite effect. The minimum value of n is 5.
- CLUSTER=name
-
specifies a name for the variable in the OUTSEED= and
OUT= data sets that indicates cluster membership.
The default name for this variable is CLUSTER.
- CONVERGE=c
- CONV=c
-
specifies the convergence criterion.
Any nonnegative value is allowed.
The default value is 0.0001 for all values of p if LEAST=p
is explicitly specified; otherwise, the default value is 0.02.
Iterations stop when the maximum relative change in the
cluster seeds is less than or equal to the convergence
criterion and additional conditions on the homotopy
parameter, if any, are satisfied (see the HP= option).
The relative change in a cluster seed is the distance
between the old seed and the new seed divided by a scaling factor.
If you do not specify the LEAST= option, the scaling factor
is the minimum distance between the initial seeds.
If you specify the LEAST= option, the scaling factor is an
L1 scale estimate and is recomputed on each iteration.
Specify the CONVERGE= option only if you specify
a MAXITER= value greater than 1.
- DATA=SAS-data-set
-
specifies the input data set containing observations to be clustered.
If you omit the DATA= option, the most
recently created SAS data set is used.
The data must be coordinates, not
distances, similarities, or correlations.
- DELETE=n
-
deletes cluster seeds to which n or
fewer observations are assigned.
Deletion occurs after processing for the DRIFT option is completed
and after each iteration specified by the MAXITER= option.
Cluster seeds are not deleted after the final assignment
of observations to clusters, so in rare cases a
final cluster may not have more than n members.
The DELETE= option is ineffective if you
specify MAXITER=0 and do not specify the DRIFT option.
By default, no cluster seeds are deleted.
- DISTANCE | DIST
-
computes distances between the cluster means.
- DRIFT
-
executes the second of the four steps described in
the section "Background".
After initial seed selection, each observation
is assigned to the cluster with the nearest seed.
After an observation is processed, the seed of the cluster
to which it is assigned is recalculated as the mean
of the observations currently assigned to the cluster.
Thus, the cluster seeds drift about rather than
remaining fixed for the duration of the pass.
- HC=c
- HP=p1 <p2>
-
pertains to the homotopy parameter for LEAST=p, where 1<p<2.
You should specify these options only if you encounter
convergence problems using the default values.
For 1<p<2, PROC FASTCLUS tries to optimize a
perturbed variant of the Lp clustering
criterion (Gonin and Money 1989, pp. 5 -6).
When the homotopy parameter is 0, the optimization
criterion is equivalent to the clustering criterion.
For a large homotopy parameter, the optimization
criterion approaches the least-squares criterion
and is, therefore, easy to optimize.
Beginning with a large homotopy parameter, PROC
FASTCLUS gradually decreases it by a factor in the
range [0.01,0.5] over the course of the iterations.
When both the homotopy parameter and the
convergence measure are sufficiently small,
the optimization process is declared to have converged.
If the initial homotopy parameter is too
large or if it is decreased too slowly, the
optimization may require many iterations.
If the initial homotopy parameter is too small or if it is
decreased too quickly, convergence to a local optimum is likely.
- HC=c
- specifies the criterion for updating the homotopy parameter.
The homotopy parameter is updated when the maximum relative
change in the cluster seeds is less than or equal to c.
The default is the minimum of 0.01 and 100
times the value of the CONVERGE= option.
- HP=p1
- specifies p1 as the initial value of the homotopy parameter.
The default is 0.05 if the modified Ekblom-Newton
method is used; otherwise, it is 0.25.
- HP=p1 p2
- also specifies p2 as the minimum value for the
homotopy parameter, which must be reached for convergence.
The default is the minimum of p1 and 0.01
times the value of the CONVERGE= option.
- IMPUTE
-
requests imputation of missing values after the
final assignment of observations to clusters.
If an observation has a missing value for a variable
used in the cluster analysis, the missing value is
replaced by the corresponding value in the cluster
seed to which the observation is assigned.
If the observation is not assigned to a
cluster, missing values are not replaced.
If you specify the IMPUTE option, the imputed values are not
used in computing cluster statistics.
If you also request an OUT= data set,
it contains the imputed values.
- INSTAT=SAS-data-set
-
reads a SAS data set previously created by the
FASTCLUS procedure using the OUTSTAT= option. If you specify the
INSTAT= option,
no clustering iterations are performed and no output is
displayed. Only cluster assignment and imputation are performed as an
OUT= data set is created.
- IRLS
-
causes PROC FASTCLUS to use an
iteratively reweighted least-squares method
instead of the modified Ekblom-Newton method. If you specify the IRLS
option, you must also specify LEAST=p, where 1<p<2.
Use the IRLS option only if you encounter
convergence problems with the default method.
- LEAST=p | MAX
- L=p | MAX
-
causes PROC FASTCLUS to optimize an Lp criterion, where
 (Spath 1985, pp. 62 -63).
Infinity is indicated by LEAST=MAX.
The value of this clustering criterion is displayed in the iteration history.
(Spath 1985, pp. 62 -63).
Infinity is indicated by LEAST=MAX.
The value of this clustering criterion is displayed in the iteration history.
If you do not specify the LEAST= option, PROC FASTCLUS
uses the least-squares (L2) criterion.
However, the default number of iterations is
only 1 if you omit the LEAST= option, so the
optimization of the criterion is generally not completed.
If you specify the LEAST= option, the maximum
number of iterations is increased to allow
the optimization process a chance to converge.
See the
MAXITER= option.
Specifying the LEAST= option also changes the
default convergence criterion from 0.02 to 0.0001.
See the CONVERGE= option.
When LEAST=2, PROC FASTCLUS tries to minimize
the root mean square difference between the
data and the corresponding cluster means.
When LEAST=1, PROC FASTCLUS tries to minimize
the mean absolute difference between the data
and the corresponding cluster medians.
When LEAST=MAX, PROC FASTCLUS tries to minimize
the maximum absolute difference between the data
and the corresponding cluster midranges.
For general values of p, PROC FASTCLUS tries to minimize the
pth root of the mean of the pth powers of the absolute
differences between the data and the corresponding cluster seeds.
The divisor in the clustering criterion is either the
number of nonmissing data used in the analysis or, if there
is a WEIGHT statement, the sum of the weights corresponding
to all the nonmissing data used in the analysis (that is,
an observation with n nonmissing data contributes n
times the observation weight to the divisor).
The divisor is not adjusted for degrees of freedom.
The method for updating cluster seeds during iteration depends
on the LEAST= option, as follows (Gonin and Money 1989).
|
LEAST=p
|
Algorithm for Computing Cluster Seeds
|
| p=1 | bin sort for median |
| 1<p<2 | modified Merle-Spath if you specify IRLS, |
| | otherwise modified Ekblom-Newton |
| p=2 | arithmetic mean |
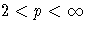 | Newton |
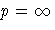 | midrange |
During the final pass, a modified Merle-Spath step is taken to
compute the cluster centers for  or
or 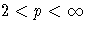 .
If you specify the LEAST=p option with a value other than 2,
PROC FASTCLUS computes pooled scale estimates analogous to the
root mean square standard deviation but based on pth power
deviations instead of squared deviations.
.
If you specify the LEAST=p option with a value other than 2,
PROC FASTCLUS computes pooled scale estimates analogous to the
root mean square standard deviation but based on pth power
deviations instead of squared deviations.
|
LEAST=p
|
Scale Estimate
|
| p=1 | mean absolute deviation |
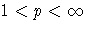 | root mean pth-power absolute deviation |
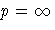 | maximum absolute deviation |
The divisors for computing the mean absolute
deviation or the root mean pth-power absolute
deviation are adjusted for degrees of freedom just
like the divisors for computing standard deviations.
This adjustment can be suppressed by the VARDEF= option.
- LIST
-
lists all observations, giving the value of the ID
variable (if any), the number of the cluster to
which the observation is assigned, and the distance
between the observation and the final cluster seed.
- MAXITER=n
-
specifies the maximum number of
iterations for recomputing cluster seeds.
When the value of the MAXITER= option is greater than 0, PROC FASTCLUS
executes the third of the four steps described in
the "Background" section.
In each iteration, each observation is
assigned to the nearest seed, and the seeds
are recomputed as the means of the clusters.
The default value of the MAXITER=
option depends on the LEAST=p option.
|
LEAST=p
|
MAXITER=
|
| not specified | 1 |
| p = 1 | 20 |
| 1 < p < 1.5 | 50 |
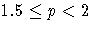 | 20 |
| p = 2 | 10 |
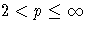 | 20 |
- MEAN=SAS-data-set
-
creates an output data set to contain the cluster
means and other statistics for each cluster.
If you want to create a permanent SAS data set,
you must specify a two-level name.
Refer to "SAS Data Files" in SAS Language Reference:
Concepts for more information on permanent data sets.
- NOMISS
-
excludes observations with missing values from the analysis.
However, if you also specify the IMPUTE option, observations
with missing values are included in the final cluster assignments.
- NOPRINT
-
suppresses the display of all output. Note that this option
temporarily disables the Output Delivery System (ODS).
For more information, see Chapter 15, "Using the Output Delivery System."
- OUT=SAS-data-set
-
creates an output data set to contain all the original
data, plus the new variables CLUSTER and DISTANCE.
Refer to "SAS Data Files" in SAS Language Reference:
Concepts for more information on permanent data sets.
- OUTITER
-
outputs information from the iteration history to the OUTSEED=
data set, including the cluster seeds at each iteration.
- OUTSEED=SAS-data-set
- OUTS=SAS-data-set
-
is another name for the MEAN= data set, provided because the
data set may contain location estimates other than means.
The MEAN= option is still accepted.
- OUTSTAT=SAS-data-set
-
creates an output data set to contain various statistics,
especially those not included in the OUTSEED= data set.
Unlike the OUTSEED= data set, the OUTSTAT=
data set is not suitable for use as a SEED=
data set in a subsequent PROC FASTCLUS step.
- RANDOM=n
-
specifies a positive integer as a starting value for the
pseudo-random number generator for use with REPLACE=RANDOM.
If you do not specify the RANDOM= option, the time of day
is used to initialize the pseudo-random number sequence.
- REPLACE=FULL | PART | NONE | RANDOM
-
specifies how seed replacement is performed.
- FULL
- requests default seed replacement as
described in the section "Background".
- PART
- requests seed replacement only when the distance
between the observation and the closest seed is
greater than the minimum distance between seeds.
- NONE
- suppresses seed replacement.
- RANDOM
- selects a simple pseudo-random sample of complete observations
as initial cluster seeds.
- SEED=SAS-data-set
-
specifies an input data set from which
initial cluster seeds are to be selected.
If you do not specify the SEED= option, initial
seeds are selected from the DATA= data set.
The SEED= data set must contain the same
variables that are used in the data analysis.
- SHORT
-
suppresses the display of the initial cluster
seeds, cluster means, and standard deviations.
- STRICT
- STRICT=s
-
prevents an observation from being assigned
to a cluster if its distance to the nearest
cluster seed exceeds the value of the STRICT= option.
If you specify the STRICT option without a numeric value,
you must also specify
the RADIUS= option, and its value is used instead.
In the OUT= data set, observations that are not assigned due
to the STRICT= option are given a negative cluster number, the absolute
value of which indicates the cluster with the nearest seed.
- SUMMARY
-
suppresses the display of the initial cluster seeds, statistics
for variables, cluster means, and standard deviations.
- VARDEF=DF | N | WDF | WEIGHT | WGT
-
specifies the divisor to be used in the
calculation of variances and covariances.
The default value is VARDEF=DF.
The possible values of the VARDEF= option and
associated divisors are as follows.
|
Value
|
|
Description
|
|
Divisor
|
| DF | | error degrees of freedom | | n-c |
| N | | number of observations | | n |
| WDF | | sum of weights DF | | 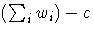 |
| WEIGHT | WGT | | sum of weights | | 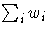 |
In the preceding definitions, c represents the number of clusters.
Copyright © 1999 by SAS Institute Inc., Cary, NC, USA. All rights reserved.