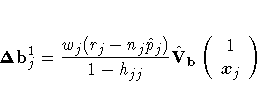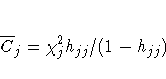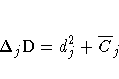Regression Diagnostics
For binary response data, regression diagnostics developed by Pregibon (1981)
can be requested by specifying the INFLUENCE option.
This section uses the following notation:
- rj, nj
- rj is the number of event responses out of nj trials for the
jth observation.
If events/trials syntax is used,
rj is the value of events and
nj is the value of trials. For single-trial
syntax, nj=1, and rj=1 if the ordered
response is 1, and rj=0 if the ordered response is 2.
- wj
- is the total weight (the product of the WEIGHT and FREQ values)
of the jth observation.
- pj
- is the probability of an event response for the jth observation
given by
 , where F(.) is the
inverse link function.
, where F(.) is the
inverse link function.
- b
- is the maximum likelihood estimate (MLE)
of
 .
.
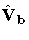
- is the estimated covariance matrix of b.

 is the estimate of pj
evaluated at b, and
is the estimate of pj
evaluated at b, and  .
.
Pregibon suggests using the index plots of several diagnostic statistics
to identify influential observations and to quantify the effects on various
aspects of the maximum likelihood fit.
In an index plot, the diagnostic statistic is plotted
against the observation number.
In general, the distributions of these diagnostic statistics are not known,
so cutoff values cannot be given for determining when the values are
large. However, the IPLOTS and INFLUENCE options provide displays of the
diagnostic values allowing visual inspection and comparison of
the values across observations. In these plots, if the model is
correctly specified and fits all observations well, then no extreme
points should appear.
The next five sections give formulas
for these diagnostic statistics.
The diagonal elements of the hat matrix are useful in detecting extreme points
in the design space where they tend to have larger values. The jth
diagonal element is
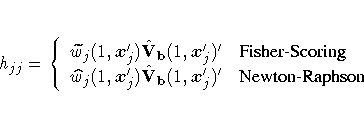
where
![\widetilde{w}_j & = & \frac{w_j n_j}{\hat{p}_j\hat{q}_j[g'(\hat{p}_j)]^2} \ \hat...
...\hat{q}_j-\hat{p}_j)g'(\hat{p}_j)]}
{ (\hat{p}_j\hat{q}_j)^2 [g'(\hat{p}_j)]^3}](images/lgseq226.gif)
g'(.) and g''(.) are the first and second derivatives of the link
function g(.), respectively.
For a binary response logit model, the hat matrix diagonal elements are

If the estimated probability is extreme
(less than 0.1 and greater than 0.9, approximately), then the hat
diagonal may be greatly reduced in value. Consequently, when an observation has
a very large or very small estimated probability, its hat diagonal value
is not a good indicator of the observation's distance from the design
space (Hosmer and Lemeshow 1989).
Pearson Residuals and Deviance Residuals
Pearson and Deviance residuals are useful in identifying observations that are
not explained well by the model. Pearson residuals are components of the
Pearson chi-square statistic and deviance residuals are components of the
deviance.
The Pearson residual for the jth
observation is

The Pearson chi-square statistic is the sum of
squares of the Pearson residuals.
The deviance residual for the jth observation is
![\hspace*{-0.5in} d_j= & \{ -\sqrt{-2w_jn_j\log(\hat{q}_j)} & {if }r_j=0 \ +-\sqr...
... ]} &
{if }0\lt r_j\lt n_j \ \sqrt{-2w_jn_j\log(\hat{p}_j)} & {if }r_j=n_j
. \](images/lgseq229.gif)
where the plus (minus) in  is used if
rj/nj is greater (less) than
is used if
rj/nj is greater (less) than
 . The deviance is the sum of squares of the
deviance residuals.
. The deviance is the sum of squares of the
deviance residuals.
DFBETAs
For each parameter estimate, the procedure calculates a DFBETA diagnostic
for each observation. The DFBETA diagnostic for an observation
is the standardized difference in the parameter estimate
due to deleting the
observation, and it can be used to assess the effect of an individual
observation on each estimated parameter of the fitted model.
Instead of re-estimating
the parameter every time an observation is deleted, PROC LOGISTIC uses
the one-step estimate. See the section "Predicted Probability of an Event for Classification". For the jth observation,
the DFBETAS are given by

where  is the standard error of the ith
component of b, and
is the standard error of the ith
component of b, and
 is
the ith component of the one-step difference
is
the ith component of the one-step difference
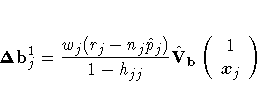
 is the approximate change (b- bj1) in
the vector of parameter estimates due to the omission of the jth
observation.
The DFBETAs are useful in detecting observations that are causing
instability in the selected coefficients.
is the approximate change (b- bj1) in
the vector of parameter estimates due to the omission of the jth
observation.
The DFBETAs are useful in detecting observations that are causing
instability in the selected coefficients.
C and CBAR
C and CBAR are confidence interval displacement diagnostics that
provide scalar
measures of the influence of individual observations on b.
These diagnostics are based on the same idea
as the Cook distance in linear regression theory, and by using
the one-step estimate, C and CBAR for the jth observation
are computed as

and
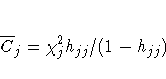
respectively.
Typically, to use these statistics, you plot them against an
index (as the IPLOT option does)
and look for
outliers.
DIFDEV and DIFCHISQ
DIFDEV and DIFCHISQ are diagnostics for detecting ill-fitted
observations; in other words, observations that contribute heavily to
the
disagreement between the data and the predicted values of the fitted
model. DIFDEV is the change in the deviance due to deleting an
individual observation while DIFCHISQ is the change in the Pearson
chi-square statistic for the same deletion. By using the one-step
estimate,
DIFDEV and DIFCHISQ for the jth observation are computed as
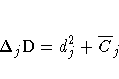
and

Copyright © 1999 by SAS Institute Inc., Cary, NC, USA. All rights reserved.
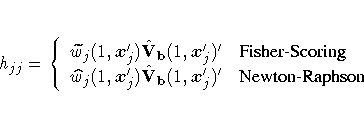
![\widetilde{w}_j & = & \frac{w_j n_j}{\hat{p}_j\hat{q}_j[g'(\hat{p}_j)]^2} \ \hat...
...\hat{q}_j-\hat{p}_j)g'(\hat{p}_j)]}
{ (\hat{p}_j\hat{q}_j)^2 [g'(\hat{p}_j)]^3}](images/lgseq226.gif)


![\hspace*{-0.5in} d_j= & \{ -\sqrt{-2w_jn_j\log(\hat{q}_j)} & {if }r_j=0 \ +-\sqr...
... ]} &
{if }0\lt r_j\lt n_j \ \sqrt{-2w_jn_j\log(\hat{p}_j)} & {if }r_j=n_j
. \](images/lgseq229.gif)