Chapter Contents
Previous
Next
|
Chapter Contents |
Previous |
Next |
| The MULTTEST Procedure |
PROC MULTTEST offers p-value adjustments using Bonferroni, Sidak, Bootstrap resampling, and Permutation resampling, all with single-step or stepdown versions. In addition, Hochberg's (1988) and Benjamini and Hochberg's (1995) step-up methods are offered. The Bonferroni and Sidak methods are calculated from the permutation distributions when exact permutation tests are used with CA or PETO tests.
All methods but the resampling methods are calculated using simple functions of the raw p-values or marginal permutation distributions; the permutation and bootstrap adjustments require the raw data. Because the resampling techniques incorporate distributional and correlational structures, they tend to be less conservative than the other methods.
When a resampling (bootstrap or permutation) method is used with only one test, the adjusted p-value is the bootstrap or permutation p-value for that test, with no adjustment for multiplicity, as described by Westfall and Soper (1994).
If the unadjusted p-values are computed using exact permutation
distributions, then the Bonferroni adjustment for pr is p1*+ ... + pR*, where pj* is the largest p-value from the
permutation distribution of test j satisfying ![]() , or
0 if all permutational p-values of test j are greater than
pr. These adjustments are much less conservative than the
ordinary Bonferroni adjustments because they incorporate the
discrete distributional characteristics. However, they remain
conservative in that they do not incorporate correlation structures
between multiple contrasts and multiple variables (Westfall and
Wolfinger 1997).
, or
0 if all permutational p-values of test j are greater than
pr. These adjustments are much less conservative than the
ordinary Bonferroni adjustments because they incorporate the
discrete distributional characteristics. However, they remain
conservative in that they do not incorporate correlation structures
between multiple contrasts and multiple variables (Westfall and
Wolfinger 1997).
If the unadjusted p-values are computed using exact permutation distributions, then the Sidak adjustment for pr is 1-(1-p1*) ... (1- pR*), where the pj* are as described previously. These adjustments are less conservative than the corresponding Bonferroni adjustments, but they do not incorporate correlation structures between multiple contrasts and multiple variables (Westfall and Wolfinger 1997).
In the case of continuous data, the pooling of the groups is not likely to recreate the shape of the null hypothesis distribution, since the pooled data are likely to be multimodal. For this reason, PROC MULTTEST automatically mean-centers all continuous variables prior to resampling. Such mean-centering is akin to resampling residuals in a regression analysis, as discussed by Freedman (1981). You can specify the NOCENTER option if you do not want to center the data. (In most situations, it does not seem to make much difference whether or not you center the data.)
The bootstrap method explicitly incorporates all sources of correlation, from both the multiple contrasts and the multivariate structure. The adjusted p-values incorporate all correlations and distributional characteristics.
The permutation method explicitly incorporates all sources of correlation, from both the multiple contrasts and the multivariate structure. The adjusted p-values incorporate all correlations and distributional characteristics.
Suppose the base test p-values are ordered as p1 < p2 < ... < pR. The Bonferroni stepdown p-values s1, ... ,sR are obtained from
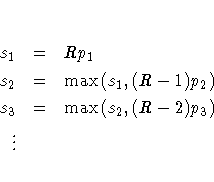
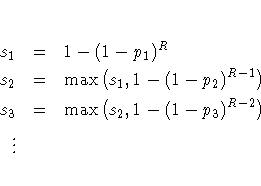
Stepdown Bonferroni adjustments using exact tests are defined as
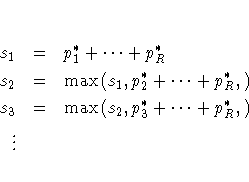
Stepdown Sidak adjustments for exact tests are defined analogously by substituting 1-(1-pj*) ... (1-pR*) for pj* + ... + pR*.
The resampling-style stepdown method is analogous to the preceding stepdown methods; the most extreme p-value is adjusted according to all R tests, the second-most extreme p-value is adjusted according to (R - 1) tests, and so on. The difference is that all correlational and distributional characteristics are incorporated when you use resampling methods. More specifically, assuming the same ordering of p-values as discussed previously, the resampling-style stepdown adjusted p-value for test r is the probability that the minimum pseudo-p-value of tests r, ... ,R is less than or equal to pr.
This probability is evaluated using Monte Carlo, as are the previously described resampling-style adjusted p-values. In fact, the computations for stepdown adjusted p-values are essentially no more time-consuming than the computations for the nonstepdown adjusted p-values. After Monte Carlo, the stepdown adjusted p-values are corrected to ensure monotonicity; this correction leaves the first adjusted p-values alone, then corrects the remaining ones as needed. The stepdown method approximately controls the familywise error rate, and it is described in more detail by Westfall and Young (1993).
The Hochberg adjusted p-values are defined in reverse order as the stepdown Bonferroni:
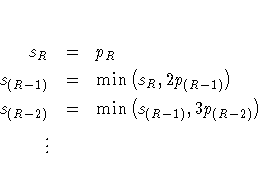
The FDR adjusted p-values are defined in step-up fashion, like the Hochberg adjustments, but with less conservative multipliers:
![s_R & = & p_R \ s_{(R-1)} & = & \min ( s_R , [R/(R-1)] p_{(R-1)} ) \ s_{(R-2)} & = & \min ( s_{(R-1)} , [R/(R-2)] p_{(R-2)} ) \ \vdots & &](images/mlteq43.gif)
|
Chapter Contents |
Previous |
Next |
Top |
Copyright © 1999 by SAS Institute Inc., Cary, NC, USA. All rights reserved.