Chapter Contents
Previous
Next
|
Chapter Contents |
Previous |
Next |
| Details of the OPTEX Procedure |
| See OPTEX10 in the SAS/QC Sample Library |
The BLOCKS statement finds a design that maximizes the determinant |X'AX| of the treatment information matrix, where A depends on the block or covariate model. Alternatively, you can directly specify the matrix A to find the D-optimal design when A is the variance-covariance matrix for the runs. You can specify the data set containing the covariance matrix with the COVAR= option in the BLOCKS statement, listing the variables corresponding to the columns of the covariance matrix in the VAR= option. If you specify n variables in the VAR= option, the values of these variables in the first n observations in the data set will be used to define A.
For example, suppose you want to compare the effects of seven different fertilizers on crop yield, using seven long, narrow blocks of four plots each, as depicted in Figure 24.1.
Figure 24.1: Block Structure for Neighbor Balance
In this case, it is reasonable to conjecture that closer plots within each block are more correlated. In particular, suppose that the plots are autocorrelated, so that the correlation matrix for the four plots in each block is of the form
![R & = & [1 & \rho & \rho^2 & \rho^3 \ \rho & 1 & \rho & \rho^2 \ \rho^2 & \rho & 1 & \rho \ \rho^3 & \rho^2 & \rho & 1
]](images/opteq8.gif)
where ![]() . If there is also an overall fixed effect
due to blocks, the information matrix for the effect of fertilizer
has the form X'AX, where
. If there is also an overall fixed effect
due to blocks, the information matrix for the effect of fertilizer
has the form X'AX, where
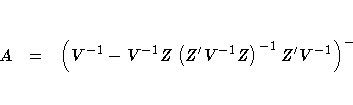
The following code uses the SAS/IML matrix language to
construct A using ![]() and saves it in a data set named A:
and saves it in a data set named A:
proc iml;
blks = int(((1:28)`-1)/4) + 1;
z = j(28,1) || designf(blks);
r = toeplitz(0.1**(0:3));
v = r;
do i = 2 to 7; v = block(v,r); end;
iv = inv(v);
a = ginv(iv-iv*z*inv(z`*iv*z)*z`*iv);
create A from a;
append from a;
quit;
Note that the data set is created with variables named COL1, COL2, ...,
COL28, by default.
To find an allocation of fertilizers to plots that is optimal for detecting the fertilizer effect in the presence of this autocorrelation, simply specify a one-way model for the treatment effects and specify the data set A as the covariance matrix for the runs with the COVAR= option in the BLOCKS statement, as follows:
data fert; do f = 1 to 7; output; end;
proc optex data=fert seed=56672 coding=orth;
class f;
model f;
blocks covar=A
var=(col1-col28);
output out=nbd;
run;
The SAS/IML matrix language also provides a convenient way of listing the
design.
proc iml;
use nbd; Read in the selected levels
read all var {f}; of fertilizer
nbd = shape(f,7,4); Reshape them into 7 4-run
print nbd [format=2.]; blocks and print.
The resulting design is shown in Output 24.9.1. Note that it is not only
a balanced incomplete block design, but it is also balanced for first
neighbors; that is, every pair of treatments occur equally often
on horizontally adjacent plots.
Output 24.9.1: Neighbor-Balanced BIBD for v=b=7, k=4, Found by
Optimal Blocking
| ||||||||||||||||||||||||||||||||
|
Chapter Contents |
Previous |
Next |
Top |
Copyright © 1999 by SAS Institute Inc., Cary, NC, USA. All rights reserved.