Chapter Contents
Previous
Next
|
Chapter Contents |
Previous |
Next |
| The ACECLUS Procedure |
The iris data published by Fisher (1936) have been widely used for examples in discriminant analysis and cluster analysis. The sepal length, sepal width, petal length, and petal width are measured in millimeters on fifty iris specimens from each of three species, Iris setosa, I. versicolor, and I. virginica. Mezzich and Solomon (1980) discuss a variety of cluster analyses of the iris data.
In this example PROC ACECLUS is used to transform the data, and the clustering is performed by PROC FASTCLUS. Compare this with the example in Chapter 27, "The FASTCLUS Procedure." The results from the FREQ procedure display fewer misclassifications when PROC ACECLUS is used. The following statements produce Output 16.1.1 through Output 16.1.5.
proc format;
value specname
1='Setosa '
2='Versicolor'
3='Virginica ';
run;
data iris;
title 'Fisher (1936) Iris Data';
input SepalLength SepalWidth PetalLength PetalWidth Species @@;
format Species specname.;
label SepalLength='Sepal Length in mm.'
SepalWidth ='Sepal Width in mm.'
PetalLength='Petal Length in mm.'
PetalWidth ='Petal Width in mm.';
symbol = put(species, specname10.);
datalines;
50 33 14 02 1 64 28 56 22 3 65 28 46 15 2 67 31 56 24 3
63 28 51 15 3 46 34 14 03 1 69 31 51 23 3 62 22 45 15 2
59 32 48 18 2 46 36 10 02 1 61 30 46 14 2 60 27 51 16 2
65 30 52 20 3 56 25 39 11 2 65 30 55 18 3 58 27 51 19 3
68 32 59 23 3 51 33 17 05 1 57 28 45 13 2 62 34 54 23 3
77 38 67 22 3 63 33 47 16 2 67 33 57 25 3 76 30 66 21 3
49 25 45 17 3 55 35 13 02 1 67 30 52 23 3 70 32 47 14 2
64 32 45 15 2 61 28 40 13 2 48 31 16 02 1 59 30 51 18 3
55 24 38 11 2 63 25 50 19 3 64 32 53 23 3 52 34 14 02 1
49 36 14 01 1 54 30 45 15 2 79 38 64 20 3 44 32 13 02 1
67 33 57 21 3 50 35 16 06 1 58 26 40 12 2 44 30 13 02 1
77 28 67 20 3 63 27 49 18 3 47 32 16 02 1 55 26 44 12 2
50 23 33 10 2 72 32 60 18 3 48 30 14 03 1 51 38 16 02 1
61 30 49 18 3 48 34 19 02 1 50 30 16 02 1 50 32 12 02 1
61 26 56 14 3 64 28 56 21 3 43 30 11 01 1 58 40 12 02 1
51 38 19 04 1 67 31 44 14 2 62 28 48 18 3 49 30 14 02 1
51 35 14 02 1 56 30 45 15 2 58 27 41 10 2 50 34 16 04 1
46 32 14 02 1 60 29 45 15 2 57 26 35 10 2 57 44 15 04 1
50 36 14 02 1 77 30 61 23 3 63 34 56 24 3 58 27 51 19 3
57 29 42 13 2 72 30 58 16 3 54 34 15 04 1 52 41 15 01 1
71 30 59 21 3 64 31 55 18 3 60 30 48 18 3 63 29 56 18 3
49 24 33 10 2 56 27 42 13 2 57 30 42 12 2 55 42 14 02 1
49 31 15 02 1 77 26 69 23 3 60 22 50 15 3 54 39 17 04 1
66 29 46 13 2 52 27 39 14 2 60 34 45 16 2 50 34 15 02 1
44 29 14 02 1 50 20 35 10 2 55 24 37 10 2 58 27 39 12 2
47 32 13 02 1 46 31 15 02 1 69 32 57 23 3 62 29 43 13 2
74 28 61 19 3 59 30 42 15 2 51 34 15 02 1 50 35 13 03 1
56 28 49 20 3 60 22 40 10 2 73 29 63 18 3 67 25 58 18 3
49 31 15 01 1 67 31 47 15 2 63 23 44 13 2 54 37 15 02 1
56 30 41 13 2 63 25 49 15 2 61 28 47 12 2 64 29 43 13 2
51 25 30 11 2 57 28 41 13 2 65 30 58 22 3 69 31 54 21 3
54 39 13 04 1 51 35 14 03 1 72 36 61 25 3 65 32 51 20 3
61 29 47 14 2 56 29 36 13 2 69 31 49 15 2 64 27 53 19 3
68 30 55 21 3 55 25 40 13 2 48 34 16 02 1 48 30 14 01 1
45 23 13 03 1 57 25 50 20 3 57 38 17 03 1 51 38 15 03 1
55 23 40 13 2 66 30 44 14 2 68 28 48 14 2 54 34 17 02 1
51 37 15 04 1 52 35 15 02 1 58 28 51 24 3 67 30 50 17 2
63 33 60 25 3 53 37 15 02 1
;
proc aceclus data=iris out=ace p=.02 outstat=score;
var SepalLength SepalWidth PetalLength PetalWidth ;
run;
legend1 frame cframe=ligr cborder=black position=center
value=(justify=center);
axis1 label=(angle=90 rotate=0) minor=none;
axis2 minor=none;
proc gplot data=ace;
plot can2*can1=Species/
frame cframe=ligr legend=legend1 vaxis=axis1 haxis=axis2;
format Species specname. ;
run;
proc fastclus data=ace maxc=3 maxiter=10 conv=0 out=clus;
var can:;
run;
proc freq;
tables cluster*Species;
run;
Output 16.1.1: Using PROC ACECLUS to Transform Fisher's Iris Data
|
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Output 16.1.2: Eigenvalues, Raw Canonical Coefficients, and Standardized Canonical Coefficients
|
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
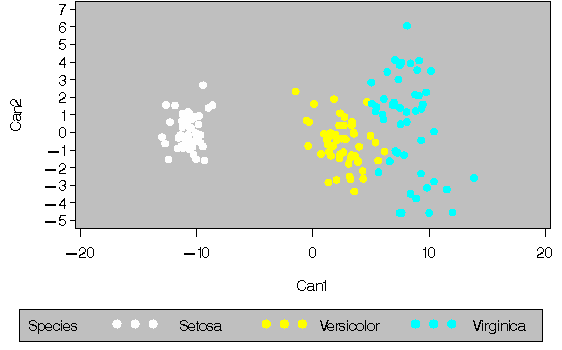
|
Output 16.1.4: Clustering of Transformed Iris Data: Partial Output from PROC FASTCLUS
|
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Output 16.1.5: Crosstabulation of Cluster by Species for Fisher's Iris Data: PROC FREQ
|
| |||||||||||||||||||||||||||||||||||||
|
Chapter Contents |
Previous |
Next |
Top |
Copyright © 1999 by SAS Institute Inc., Cary, NC, USA. All rights reserved.