PROC DISCRIM Statement
- PROC DISCRIM < options > ;
This statement invokes the DISCRIM procedure.
You can specify the following options in the PROC DISCRIM statement.
|
Tasks
|
|
Options
|
| Specify Input Data Set | | DATA= |
| | | | TESTDATA= |
| Specify Output Data Set | | OUTSTAT= |
| | | | OUT= |
| | | | OUTCROSS= |
| | | | OUTD= |
| | | | TESTOUT= |
| | | | TESTOUTD= |
| Discriminant Analysis | | METHOD= |
| | | | POOL= |
| | | | SLPOOL= |
| Nonparametric Methods | | K= |
| | | | R= |
| | | | KERNEL= |
| | | | METRIC= |
|
Tasks
|
|
Options
|
| Classification Rule | | THRESHOLD= |
| Determine Singularity | | SINGULAR= |
| Canonical Discriminant Analysis | | CANONICAL |
| | | | CANPREFIX= |
| | | | NCAN= |
| Resubstitution Classification | | LIST |
| | | | LISTERR |
| | | | NOCLASSIFY |
| Cross Validation Classification | | CROSSLIST |
| | | | CROSSLISTERR |
| | | | CROSSVALIDATE |
| Test Data Classification | | TESTLIST |
| | | | TESTLISTERR |
| Estimate Error Rate | | POSTERR |
| Control Displayed Output | | |
| | Correlations | | BCORR |
| | | | PCORR |
| | | | TCORR |
| | | | WCORR |
| | Covariances | | BCOV |
| | | | PCOV |
| | | | TCOV |
| | | | WCOV |
| | SSCP Matrix | | BSSCP |
| | | | PSSCP |
| | | | TSSCP |
| | | | WSSCP |
| | Miscellaneous | | ALL |
| | | | ANOVA |
| | | | DISTANCE |
| | | | MANOVA |
| | | | SIMPLE |
| | | | STDMEAN |
| | Suppress output | | NOPRINT |
| | | | SHORT |
- ALL
-
activates all options that control displayed output.
When the derived classification criterion is used to classify
observations, the ALL option also activates the
POSTERR option.
- ANOVA
-
displays univariate statistics for testing the hypothesis that
the class means are equal in the population for each variable.
- BCORR
-
displays between-class correlations.
- BCOV
-
displays between-class covariances.
The between-class covariance matrix equals the between-class
SSCP matrix divided by n(c-1)/c, where n is the number
of observations and c is the number of classes.
You should interpret the between-class covariances
in comparison with the total-sample and within-class
covariances, not as formal estimates of population parameters.
- BSSCP
-
displays the between-class SSCP matrix.
- CANONICAL
- CAN
-
performs canonical discriminant analysis.
- CANPREFIX=name
-
specifies a prefix for naming the canonical variables.
By default, the names are Can1, Can2, ... , Cann.
If you specify CANPREFIX=ABC, the components
are named ABC1, ABC2, ABC3, and so on.
The number of characters in the prefix, plus
the number of digits required to designate the
canonical variables, should not exceed 32.
The prefix is truncated if the combined length exceeds 32.
The CANONICAL option is activated when you specify either
the NCAN= or the CANPREFIX= option.
A discriminant criterion is always derived in PROC DISCRIM.
If you want canonical discriminant analysis without the
use of discriminant criteria, you should use PROC CANDISC.
- CROSSLIST
-
displays the cross validation classification
results for each observation.
- CROSSLISTERR
-
displays the cross validation classification
results for misclassified observations only.
- CROSSVALIDATE
-
specifies the cross validation
classification of the input DATA= data set.
When a parametric method is used, PROC DISCRIM classifies each
observation in the DATA= data set using a discriminant
function computed from the other observations in the
DATA= data set, excluding the observation being classified.
When a nonparametric method is used, the covariance matrices used
to compute the distances are based on all observations in the
data set and do not exclude the observation being classified.
However, the observation being classified is excluded
from the nonparametric density estimation (if you specify the R=
option) or the k nearest neighbors
(if you specify the K= option) of that observation.
The CROSSVALIDATE option is set when you specify the CROSSLIST,
CROSSLISTERR, or OUTCROSS= option.
- DATA=SAS-data-set
-
specifies the data set to be analyzed.
The data set can be an ordinary SAS data set or one of several
specially structured data sets created by SAS/STAT procedures.
These specially structured data sets include
TYPE=CORR, TYPE=COV, TYPE=CSSCP, TYPE=SSCP,
TYPE=LINEAR, TYPE=QUAD, and TYPE=MIXED.
The input data set must be an ordinary
SAS data set if you specify METHOD=NPAR.
If you omit the DATA= option, the procedure
uses the most recently created SAS data set.
- DISTANCE
- MAHALANOBIS
displays the squared Mahalanobis distances between the group
means, F statistics, and the corresponding probabilities of
greater Mahalanobis squared distances between the group means.
The squared distances are based on the
specification of the POOL=
and METRIC= options.
- K=k
-
specifies a k value for the k-nearest-neighbor rule.
An observation x is classified into a group based on
the information from the k nearest neighbors of x.
Do not specify both the K= and R= options.
- KERNEL=BIWEIGHT | BIW
- KERNEL=EPANECHNIKOV | EPA
- KERNEL=NORMAL | NOR
- KERNEL=TRIWEIGHT | TRI
- KERNEL=UNIFORM | UNI
-
specifies a kernel density to estimate the group-specific densities.
You can specify the KERNEL= option only when the
R= option is specified.
The default is KERNEL=UNIFORM.
- LIST
-
displays the resubstitution classification
results for each observation.
You can specify this option only when the
input data set is an ordinary SAS data set.
- LISTERR
-
displays the resubstitution classification
results for misclassified observations only.
You can specify this option only when the
input data set is an ordinary SAS data set.
- MANOVA
-
displays multivariate statistics for testing the hypothesis
that the class means are equal in the population.
- METHOD=NORMAL | NPAR
-
determines the method to use in
deriving the classification criterion.
When you specify METHOD=NORMAL, a parametric method based
on a multivariate normal distribution within each class is
used to derive a linear or quadratic discriminant function.
The default is METHOD=NORMAL.
When you specify METHOD=NPAR, a nonparametric method
is used and you must also specify either the
K= or R= option.
- METRIC=DIAGONAL | FULL | IDENTITY
-
specifies the metric in which the computations
of squared distances are performed.
If you specify METRIC=FULL, PROC DISCRIM uses either the pooled covariance matrix
(POOL=YES) or individual within-group covariance
matrices (POOL=NO) to compute the squared distances.
If you specify METRIC=DIAGONAL, PROC DISCRIM uses either the diagonal matrix of
the pooled covariance matrix (POOL=YES) or diagonal
matrices of individual within-group covariance
matrices (POOL=NO) to compute the squared distances.
If you specify METRIC=IDENTITY, PROC DISCRIM uses Euclidean distance.
The default is METRIC=FULL.
When you specify METHOD=NORMAL, the option METRIC=FULL is used.
- NCAN=number
-
specifies the number of canonical variables to compute.
The value of number must be less than
or equal to the number of variables.
If you specify the option NCAN=0, the procedure
displays the canonical correlations but not the
canonical coefficients, structures, or means.
Let v be the number of variables in the VAR
statement and c be the number of classes.
If you omit the NCAN= option, only
min(v, c-1) canonical variables are generated.
If you request an output data set (OUT=, OUTCROSS=, TESTOUT=),
v canonical variables are generated.
In this case, the last v-(c-1)
canonical variables have missing values.
The CANONICAL option is activated when you specify either
the NCAN= or the CANPREFIX= option.
A discriminant criterion is always derived in PROC DISCRIM.
If you want canonical discriminant analysis without the
use of discriminant criterion, you should use PROC CANDISC.
- NOCLASSIFY
-
suppresses the resubstitution
classification of the input DATA= data set.
You can specify this option only when the
input data set is an ordinary SAS data set.
- NOPRINT
-
suppresses the normal display of results. Note that this option
temporarily disables the Output Delivery System
(ODS); see Chapter 15, "Using the Output Delivery System," for more information.
- OUT=SAS-data-set
-
creates an output SAS data set containing all the data from the
DATA= data set, plus the posterior probabilities and the class
into which each observation is classified by resubstitution.
When you specify the CANONICAL option, the data set also
contains new variables with canonical variable scores.
See the "OUT= Data Set" section.
- OUTCROSS=SAS-data-set
-
creates an output SAS data set containing all the data from the
DATA= data set, plus the posterior probabilities and the class
into which each observation is classified by cross validation.
When you specify the CANONICAL option, the data set also
contains new variables with canonical variable scores.
See the "OUT= Data Set" section.
- OUTD=SAS-data-set
-
creates an output SAS data set containing all the
data from the DATA= data set, plus the group-specific
density estimates for each observation.
See the "OUT= Data Set" section.
- OUTSTAT=SAS-data-set
-
creates an output SAS data set containing various statistics
such as means, standard deviations, and correlations.
When the input data set is an ordinary SAS data set or when
TYPE=CORR, TYPE=COV, TYPE=CSSCP, or TYPE=SSCP, this
option can be used to generate discriminant statistics.
When you specify the CANONICAL option, canonical correlations,
canonical structures, canonical coefficients, and means of
canonical variables for each class are included in the data set.
If you specify METHOD=NORMAL, the output data set also
includes coefficients of the discriminant functions,
and the output data set is TYPE=LINEAR (POOL=YES),
TYPE=QUAD (POOL=NO), or TYPE=MIXED (POOL=TEST).
If you specify METHOD=NPAR, this output data set is TYPE=CORR.
This data set also holds calibration information
that can be used to classify new observations.
See the "Saving and Using Calibration Information" section and the "OUT= Data Set" section.
- PCORR
-
displays pooled within-class correlations.
- PCOV
-
displays pooled within-class covariances.
- POOL=NO | TEST | YES
-
determines whether the pooled or within-group covariance
matrix is the basis of the measure of the squared distance.
If you specify POOL=YES, PROC DISCRIM uses the pooled covariance matrix in
calculating the (generalized) squared distances.
Linear discriminant functions are computed.
If you specify POOL=NO, the procedure uses the individual within-group
covariance matrices in calculating the distances.
Quadratic discriminant functions are computed.
The default is POOL=YES.
When you specify METHOD=NORMAL, the option POOL=TEST requests
Bartlett's modification of the likelihood ratio
test (Morrison 1976; Anderson 1984) of the
homogeneity of the within-group covariance matrices.
The test is unbiased (Perlman 1980).
However, it is not robust to nonnormality.
If the test statistic is significant at the level specified by
the SLPOOL= option, the within-group covariance matrices are used.
Otherwise, the pooled covariance matrix is used.
The discriminant function coefficients are displayed
only when the pooled covariance matrix is used.
- POSTERR
-
displays the posterior probability error-rate estimates of the
classification criterion based on the classification results.
- PSSCP
-
displays the pooled within-class corrected SSCP matrix.
- R=r
-
specifies a radius r value for kernel density estimation.
With uniform, Epanechnikov, biweight, or triweight
kernels, an observation x is classified into a
group based on the information from observations y
in the training set within the radius r of x,
that is, the group t observations y with squared
distance
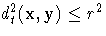 .When a normal kernel is used, the classification
of an observation x is based on the
information of the estimated group-specific
densities from all observations in the training set.
The matrix r2 Vt is used as the group t covariance
matrix in the normal-kernel density, where Vt
is the matrix used in calculating the squared distances.
Do not specify both the K= and R= options.
For more information on selecting r, see
the "Nonparametric Methods" section.
.When a normal kernel is used, the classification
of an observation x is based on the
information of the estimated group-specific
densities from all observations in the training set.
The matrix r2 Vt is used as the group t covariance
matrix in the normal-kernel density, where Vt
is the matrix used in calculating the squared distances.
Do not specify both the K= and R= options.
For more information on selecting r, see
the "Nonparametric Methods" section.
- SHORT
-
suppresses the display of certain items in the default output.
If you specify METHOD= NORMAL, PROC DISCRIM suppresses the display of
determinants, generalized squared distances between-class
means, and discriminant function coefficients.
When you specify the CANONICAL option, PROC DISCRIM suppresses
the display of canonical structures, canonical coefficients,
and class means on canonical variables; only tables of
canonical correlations are displayed.
- SIMPLE
-
displays simple descriptive statistics for
the total sample and within each class.
- SINGULAR=p
-
specifies the criterion for determining
the singularity of a matrix, where 0<p<1.
The default is SINGULAR=1E-8.
Let S be the total-sample correlation matrix.
If the R2 for predicting a quantitative variable
in the VAR statement from the variables preceding
it exceeds 1-p, then S is considered singular.
If S is singular, the probability levels for the
multivariate test statistics and canonical correlations are
adjusted for the number of variables with R2 exceeding 1-p.
Let St be the group t covariance matrix
and Sp be the pooled covariance matrix.
In group t, if the R2 for predicting a quantitative
variable in the VAR statement from the variables preceding
it exceeds 1-p, then St is considered singular.
Similarly, if the partial R2 for predicting a quantitative
variable in the VAR statement from the variables preceding it,
after controlling for the effect of the CLASS variable,
exceeds 1-p, then Sp is considered singular.
If PROC DISCRIM needs to compute either the inverse or the determinant
of a matrix that is considered singular, then it uses a quasi-inverse
or a quasi-determinant.
For details, see the "Quasi-Inverse" section.
- SLPOOL=p
-
specifies the significance level for the test of homogeneity.
You can specify the SLPOOL= option only when POOL=TEST is also specified.
If you specify POOL= TEST but omit the SLPOOL= option,
PROC DISCRIM uses 0.10 as the significance level for the test.
- STDMEAN
-
displays total-sample and pooled
within-class standardized class means.
- TCORR
-
displays total-sample correlations.
- TCOV
-
displays total-sample covariances.
- TESTDATA=SAS-data-set
-
names an ordinary SAS data set with
observations that are to be classified.
The quantitative variable names in this data
set must match those in the DATA= data set.
When you specify the TESTDATA= option, you can also specify
the TESTCLASS,
TESTFREQ, and
TESTID statements.
When you specify the TESTDATA= option, you can use the
TESTOUT=
and TESTOUTD= options to generate
classification results and group-specific density
estimates for observations in the test data set.
- TESTLIST
-
lists classification results for all
observations in the TESTDATA= data set.
- TESTLISTERR
-
lists only misclassified observations in the
TESTDATA=
data set but only if a TESTCLASS statement is also used.
- TESTOUT=SAS-data-set
-
creates an output SAS data set containing all the data from
the TESTDATA= data set, plus the posterior probabilities
and the class into which each observation is classified.
When you specify the CANONICAL option, the data set also
contains new variables with canonical variable scores.
See the "OUT= Data Set" section.
- TESTOUTD=SAS-data-set
-
creates an output SAS data set containing all
the data from the TESTDATA= data set, plus the
group-specific density estimates for each observation.
See the "OUT= Data Set" section.
- THRESHOLD=p
-
specifies the minimum acceptable posterior
probability for classification, where
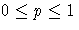 .If the largest posterior probability of group
membership is less than the THRESHOLD value,
the observation is classified into group OTHER.
The default is THRESHOLD=0.
.If the largest posterior probability of group
membership is less than the THRESHOLD value,
the observation is classified into group OTHER.
The default is THRESHOLD=0.
- TSSCP
-
displays the total-sample corrected SSCP matrix.
- WCORR
-
displays within-class correlations for each class level.
- WCOV
-
displays within-class covariances for each class level.
- WSSCP
-
displays the within-class corrected SSCP matrix for each class level.
Copyright © 1999 by SAS Institute Inc., Cary, NC, USA. All rights reserved.