Chapter Contents
Previous
Next
|
Chapter Contents |
Previous |
Next |
| The GENMOD Procedure |
This is a brief introduction to the theory of generalized linear models . See the "References" section for sources of more detailed information.
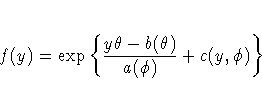
Standard theory for this type of distribution gives expressions for the mean and variance of Y.
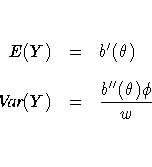
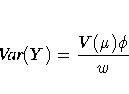
Probability distributions of the response Y in
generalized linear models are usually parameterized
in terms of the mean ![]() and dispersion parameter
and dispersion parameter
![]() instead of the natural parameter
instead of the natural parameter ![]() .The probability distributions that are available in
the GENMOD procedure are shown in the following list.
The PROC GENMOD scale parameter and
the variance of Y are also shown.
.The probability distributions that are available in
the GENMOD procedure are shown in the following list.
The PROC GENMOD scale parameter and
the variance of Y are also shown.
For the binomial distribution, the response is the binomial
proportion Y = events/ trials.
The variance function is ![]() , and the
binomial trials parameter n is regarded as a weight w.
, and the
binomial trials parameter n is regarded as a weight w.
If a weight variable is present, ![]() is replaced
with
is replaced
with ![]() , where w is the weight variable.
, where w is the weight variable.
PROC GENMOD works with a scale parameter
that is related to the exponential family dispersion
parameter ![]() instead of with
instead of with ![]() itself.
The scale parameters are related to the dispersion parameter as
shown previously with the probability distribution definitions.
Thus, the scale parameter output in the
"Analysis of Parameter Estimates" table is
related to the exponential family dispersion parameter.
If you specify a constant scale parameter with the SCALE=
option in the MODEL statement, it is also related to the
exponential family dispersion parameter in the same way.
itself.
The scale parameters are related to the dispersion parameter as
shown previously with the probability distribution definitions.
Thus, the scale parameter output in the
"Analysis of Parameter Estimates" table is
related to the exponential family dispersion parameter.
If you specify a constant scale parameter with the SCALE=
option in the MODEL statement, it is also related to the
exponential family dispersion parameter in the same way.
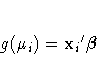
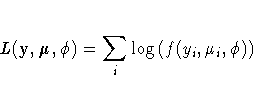
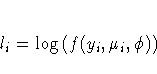
![l_i = -\frac{1}2
[ \frac{w_i(y_i-\mu_i)^2}{\phi} +
\log ( \frac{\phi}{w_i} ) + \log(2 \pi)
]](images/gmoeq72.gif)
![l_i = -\frac{1}2
[ \frac{w_i(y_i-\mu_i)^2}{y_i \mu^2 \phi} +
\log ( \frac{\phi y_i^3}{w_i} ) +
\log(2 \pi)
]](images/gmoeq73.gif)
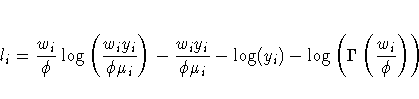
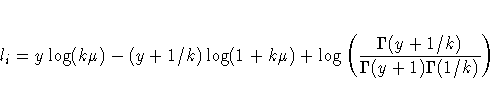
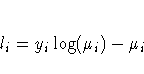
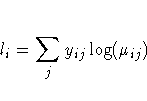
For the binomial, multinomial, and Poisson distribution, terms involving binomial coefficients or factorials of the observed counts are dropped from the computation of the log-likelihood function since they do not affect parameter estimates or their estimated covariances.
On the rth iteration, the algorithm updates the parameter
vector ![]() with
with
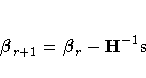
![s = [s_j]
= [ \frac{\partial L}{\partial \beta_j} ]](images/gmoeq81.gif)
![H = [{h_{ij}}]
= [ \frac{\partial^2 L}
{\partial\beta_i\partial\beta_j} ]](images/gmoeq82.gif)
In some cases, the scale parameter is estimated by maximum likelihood. In these cases, elements corresponding to the scale parameter are computed and included in s and H.
If ![]() is the linear predictor for
observation i and g is the link function, then
is the linear predictor for
observation i and g is the link function, then
![]() , so that
, so that ![]() is an estimate of the mean of the ith observation,
obtained from an estimate of the parameter vector
is an estimate of the mean of the ith observation,
obtained from an estimate of the parameter vector ![]() .
.
The gradient vector and Hessian matrix for the regression parameters are given by
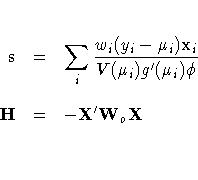
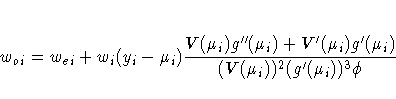
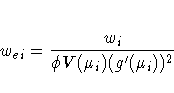
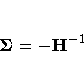
The correlation matrix
is the normalized covariance matrix.
That is, if ![]() is an element of
is an element of
![]() , then the corresponding element
of the correlation matrix is
, then the corresponding element
of the correlation matrix is ![]() ,where
,where ![]() .
.
Note that these statistics are not valid for GEE models.
If ![]() is the log-likelihood function expressed
as a function of the predicted mean values
is the log-likelihood function expressed
as a function of the predicted mean values ![]() and the vector
y of response values, then the scaled deviance
is defined by
and the vector
y of response values, then the scaled deviance
is defined by
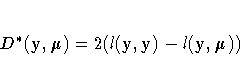
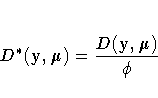
| Distribution | Deviance |
| normal | |
| Poisson | |
| binomial | |
| gamma | |
| inverse Gaussian | |
| multinomial | |
| negative binomial |
In the binomial case, yi=ri/mi, where ri is a binomial count and mi is the binomial number of trials parameter.
In the multinomial case, yij refers to the observed number of occurrences of the jth category for the ith subpopulation defined by the AGGREGATE= variable, mi is the total number in the ith subpopulation, and pij is the category probability.
Pearson's chi-square statistic is defined as
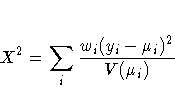
The scaled version of both of these statistics, under certain regularity conditions, has a limiting chi-square distribution, with degrees of freedom equal to the number of observations minus the number of parameters estimated. The scaled version can be used as an approximate guide to the goodness of fit of a given model. Use caution before applying these statistics to ensure that all the conditions for the asymptotic distributions hold. McCullagh and Nelder (1989) advise that differences in deviances for nested models can be better approximated by chi-square distributions than the deviances themselves.
In cases where the dispersion parameter is not known,
an estimate can be used to obtain an approximation to
the scaled deviance and Pearson's chi-square statistic.
One strategy is to fit a model that contains a sufficient
number of parameters so that all systematic variation is
removed, estimate ![]() from this model, and then use this
estimate in computing the scaled deviance of sub-models.
The deviance or Pearson's chi-square divided by its
degrees of freedom is sometimes used as an estimate
of the dispersion parameter
from this model, and then use this
estimate in computing the scaled deviance of sub-models.
The deviance or Pearson's chi-square divided by its
degrees of freedom is sometimes used as an estimate
of the dispersion parameter ![]() .For example, since the limiting chi-square distribution of the
scaled deviance
.For example, since the limiting chi-square distribution of the
scaled deviance ![]() has n-p degrees of freedom,
where n is the number of observations and p the number of
parameters, equating D*
to its mean and solving for
has n-p degrees of freedom,
where n is the number of observations and p the number of
parameters, equating D*
to its mean and solving for ![]() yields
yields ![]() .Similarly, an estimate of
.Similarly, an estimate of ![]() based on Pearson's
chi-square X2 is
based on Pearson's
chi-square X2 is ![]() .Alternatively, a maximum likelihood estimate of
.Alternatively, a maximum likelihood estimate of
![]() can be computed by the procedure, if desired.
See the discussion in
the "Type 1 Analysis" section for more on the estimation of
the dispersion parameter.
can be computed by the procedure, if desired.
See the discussion in
the "Type 1 Analysis" section for more on the estimation of
the dispersion parameter.
Otherwise, values of the SCALE and NOSCALE options and the resultant actions are displayed in the following table.
| NOSCALE | SCALE=value | Action |
| present | present | scale fixed at value |
| present | not present | scale fixed at 1 |
| not present | not present | scale estimated by ML |
| not present | present | scale estimated by ML, |
| starting point at value |
The meaning of the scale parameter displayed in the
"Analysis Of Parameter Estimates" table is different
for the Gamma distribution than for the other distributions.
The relation of the scale parameter as used by
PROC GENMOD to the exponential family dispersion
parameter ![]() is displayed in the following table.
For the binomial and Poisson distributions,
is displayed in the following table.
For the binomial and Poisson distributions,
![]() is the overdispersion parameter, as
defined in the "Overdispersion" section, which follows.
is the overdispersion parameter, as
defined in the "Overdispersion" section, which follows.
| Distribution | Scale |
| normal | |
| inverse Gaussian | |
| gamma | |
| binomial | |
| Poisson |
In the case of the negative binomial distribution, PROC GENMOD reports the "dispersion" parameter estimated by maximum likelihood. This is the negative binomial parameter k defined in the "Response Probability Distributions" section.
The SCALE= option in the MODEL statement enables
you to specify a value of ![]() for the binomial and Poisson distributions.
If you specify the SCALE=DEVIANCE option in the MODEL
statement, the procedure uses the deviance
divided by degrees of freedom as an estimate of
for the binomial and Poisson distributions.
If you specify the SCALE=DEVIANCE option in the MODEL
statement, the procedure uses the deviance
divided by degrees of freedom as an estimate of ![]() ,and all statistics are adjusted appropriately.
You can use Pearson's chi-square instead of
the deviance by specifying the SCALE=PEARSON option.
,and all statistics are adjusted appropriately.
You can use Pearson's chi-square instead of
the deviance by specifying the SCALE=PEARSON option.
The function obtained by dividing a log-likelihood function for the binomial or Poisson distribution by a dispersion parameter is not a legitimate log-likelihood function. It is an example of a quasi-likelihood function. Most of the asymptotic theory for log likelihoods also applies to quasi-likelihoods, which justifies computing standard errors and likelihood ratio statistics using quasi-likelihoods instead of proper log likelihoods. Refer to McCullagh and Nelder (1989, Chapter 9) and McCullagh (1983) for details on quasi-likelihood functions.
Although the estimate of the dispersion parameter is often used to indicate overdispersion or underdispersion, this estimate may also indicate other problems such as an incorrectly specified model or outliers in the data. You should carefully assess whether this type of model is appropriate for your data.
|
Chapter Contents |
Previous |
Next |
Top |
Copyright © 1999 by SAS Institute Inc., Cary, NC, USA. All rights reserved.