LSMEANS Statement
- LSMEANS effects < / options >
;
Least-squares means (LS-means) are computed for each
effect listed in the LSMEANS statement.
You may specify only classification effects in the LSMEANS
statement -that is, effects that contain only classification
variables.
You may also specify options to perform multiple comparisons. In
contrast to the MEANS statement, the LSMEANS statement performs
multiple comparisons on interactions as well as main effects.
LS-means are predicted population margins; that
is, they estimate the marginal means over a balanced population. In a
sense, LS-means are to unbalanced designs as class and subclass
arithmetic means are to balanced designs. Each LS-mean is computed
as L'b for a certain column vector L, where b is the vector
of parameter estimates -that is, the solution of the normal
equations.
For further information, see the section "Construction of Least-Squares Means".
Multiple effects can be specified in one LSMEANS statement, or
multiple LSMEANS statements can be used, but they must all appear
after the MODEL statement.
For example,
proc glm;
class A B;
model Y=A B A*B;
lsmeans A B A*B;
run;
LS-means are displayed for
each level of the A, B, and A*B effects.
You can specify the following options in the
LSMEANS statement after a slash:
- ADJUST=BON
-
- ADJUST=DUNNETT
- ADJUST=SCHEFFE
- ADJUST=SIDAK
- ADJUST=SIMULATE <(simoptions)>
- ADJUST=SMM | GT2
- ADJUST=TUKEY
- ADJUST=T
-
requests a multiple comparison adjustment for the p-values and
confidence limits for the differences of LS-means.
The ADJUST= option modifies the results of the TDIFF and PDIFF options;
thus, if you omit the TDIFF or PDIFF option then the ADJUST= option has
no effect.
By
default, PROC GLM analyzes all pairwise differences unless you specify
ADJUST=DUNNETT, in which case PROC GLM analyzes all differences with a
control level. The default is ADJUST=T, which really signifies no
adjustment for multiple comparisons.
The BON (Bonferroni) and SIDAK adjustments involve correction factors
described in the "Multiple Comparisons" section
and in Chapter 43, "The MULTTEST Procedure."
When you specify ADJUST=TUKEY and
your data are unbalanced, PROC GLM uses the approximation described
in Kramer (1956) and identifies the adjustment as "Tukey-Kramer" in
the results. Similarly, when you specify ADJUST=DUNNETT and the
LS-means are correlated, PROC GLM uses the
factor-analytic covariance approximation described in Hsu (1992) and
identifies the adjustment as "Dunnett-Hsu" in the results. The
preceding references also describe the SCHEFFE and SMM adjustments.
The SIMULATE adjustment computes the adjusted p-values from the simulated
distribution of the maximum or maximum absolute value of a multivariate
t random vector. The simulation estimates q, the true
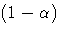 th quantile, where
th quantile, where 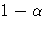 is
the confidence coefficient. The default
is
the confidence coefficient. The default 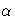 is the value of the
ALPHA= option in the PROC GLM statement or 0.05 if that option is not
specified. You can change this value with the ALPHA= option in the
LSMEANS statement.
is the value of the
ALPHA= option in the PROC GLM statement or 0.05 if that option is not
specified. You can change this value with the ALPHA= option in the
LSMEANS statement.
The number of samples for the SIMULATE adjustment is set so that the tail area
for the simulated q is within a certain accuracy radius 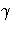 of
of 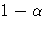 with
an accuracy confidence of
with
an accuracy confidence of 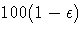 %. In equation form,
%. In equation form,

where 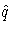 is the simulated q and F is the
true distribution function of the maximum; refer to Edwards and
Berry (1987) for details.
By default,
is the simulated q and F is the
true distribution function of the maximum; refer to Edwards and
Berry (1987) for details.
By default, 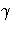 = 0.005 and
= 0.005 and 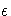 = 0.01 so that
the tail area of
= 0.01 so that
the tail area of 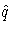 is within 0.005 of 0.95
with 99% confidence.
You can specify the following simoptions in parentheses
after the ADJUST=SIMULATE option.
is within 0.005 of 0.95
with 99% confidence.
You can specify the following simoptions in parentheses
after the ADJUST=SIMULATE option.
- ACC=value
- specifies the target accuracy radius
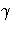 of a
of a
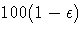 % confidence interval for the true probability
content of the estimated
% confidence interval for the true probability
content of the estimated 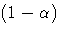 th quantile. The default value
is ACC=0.005. Note that, if you also specify the CVADJUST
simoption, then the actual accuracy radius will probably be
substantially less than this target.
th quantile. The default value
is ACC=0.005. Note that, if you also specify the CVADJUST
simoption, then the actual accuracy radius will probably be
substantially less than this target.
- CVADJUST
- specifies that the quantile should be estimated by the control variate
adjustment method of Hsu and Nelson (1998) instead of simply as the
quantile of the simulated sample. Specifying the CVADJUST option typically
has the effect of significantly reducing the accuracy radius
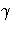 of a
of a 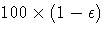 % confidence interval for the true
probability content of the estimated
% confidence interval for the true
probability content of the estimated 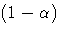 th quantile. The
control-variate-adjusted quantile estimate takes roughly twice as long
to compute, but it is typically much more accurate than the sample
quantile.
th quantile. The
control-variate-adjusted quantile estimate takes roughly twice as long
to compute, but it is typically much more accurate than the sample
quantile.
- EPS=value
- specifies the value
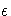 for a
for a 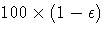 %
confidence interval for the true probability content of the estimated
%
confidence interval for the true probability content of the estimated
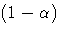 th quantile. The default value for the accuracy
confidence is 99%, corresponding to EPS=0.01.
th quantile. The default value for the accuracy
confidence is 99%, corresponding to EPS=0.01.
- NSAMP=n
- specifies the sample size for the simulation. By default, n is set
based on the values of the target accuracy radius
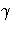 and
accuracy confidence
and
accuracy confidence 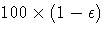 true probability content of the estimated
true probability content of the estimated 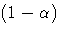 th quantile.
With the default values for
th quantile.
With the default values for 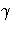 ,
, 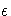 , and
, and 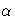 (0.005,
0.01, and 0.05, respectively), NSAMP=12604 by default.
(0.005,
0.01, and 0.05, respectively), NSAMP=12604 by default.
- REPORT
- specifies that a report on the simulation should be displayed,
including a listing of the parameters, such as
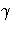 ,
, 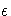 ,and
,and 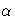 as well as an analysis of various
methods for estimating or approximating the quantile.
as well as an analysis of various
methods for estimating or approximating the quantile.
- SEED=number
- specifies a positive integer less than 231 - 1. The value of the
SEED= option is used to start the pseudo-random number generator for
the simulation. The default is a value generated from reading the
time of day from the computer's clock.
- ALPHA=p
-
specifies the level of significance p for 100(1-p)% confidence intervals.
This option is useful only if you also specify the CL option, and, optionally, the
PDIFF option. By default, p is equal to the value of the ALPHA= option
in the PROC GLM statement or 0.05 if that option is not specified,
This value is used to set the
endpoints for confidence intervals for the individual means as well as
for differences between means.
- AT variable = value
- AT (variable-list) = (value-list)
- AT MEANS
-
enables you to modify the values of the covariates used in computing
LS-means. By default, all covariate effects are set equal
to their mean values for computation of standard LS-means.
The AT option enables you to set the covariates to whatever values you
consider interesting.
For more information, see the section
"Setting Covariate Values"
- BYLEVEL
-
requests that PROC GLM process the OM data set by each level
of the LS-mean effect in question. For more details, see
the entry for the OM
option in this section.
- CL
-
requests confidence limits for the individual LS-means. If
you specify the PDIFF option, confidence limits for differences between
means are produced as well. You can control the confidence level with
the ALPHA= option. Note that, if you specify an ADJUST= option, the
confidence limits for the differences are adjusted for multiple
inference but the confidence intervals for individual means are not
adjusted.
- COV
-
includes variances and covariances of the LS-means in the output data
set specified in the OUT= option in the LSMEANS statement. Note that
this is the covariance matrix for the LS-means themselves, not the
covariance matrix for the differences between the LS-means, which is
used in the PDIFF computations.
If you omit the OUT= option, the COV option has no effect.
When you specify the COV option, you can specify
only one effect in the LSMEANS statement.
- E
-
displays the coefficients of the linear functions used to compute the LS-means.
- E=effect
-
specifies an effect in the model to use as an error term.
The procedure uses the mean square for the
effect as the error mean square when calculating estimated standard errors
(requested with the STDERR option) and probabilities
(requested with the STDERR, PDIFF, or TDIFF option).
Unless you specify STDERR, PDIFF or TDIFF,
the E= option is ignored.
By default, if you specify the STDERR, PDIFF, or TDIFF option
and do not specify the E= option, the procedure uses the error
mean square for calculating standard errors and probabilities.
- ETYPE=n
-
specifies the type (1, 2, 3, or 4, corresponding to Type I, II, III, and
IV tests, respectively) of the E= effect.
If you specify the E= option but not the ETYPE= option, the
highest type computed in the analysis is used. If you omit the E= option,
the ETYPE= option has no effect.
- NOPRINT
-
suppresses the normal display of results from the LSMEANS
statement. This option is useful when an output data set is
created with the OUT= option in the LSMEANS statement.
- OBSMARGINS
- OM
-
specifies a potentially different weighting scheme for computing
LS-means coefficients. The standard LS-means
have equal coefficients across classification effects; however,
the OM option changes these coefficients to be proportional to those
found in the input data set.
For more information, see the section
"Changing the Weighting Scheme"
The BYLEVEL option modifies the observed-margins LS-means.
Instead of computing the margins across the entire data set,
the procedure computes separate margins for each level of the LS-mean
effect in question. The resulting LS-means are actually
equal to raw means in this case.
If you specify the BYLEVEL option, it disables the AT option.
- OUT=SAS-data-set
-
creates an output data set that contains the values,
standard errors, and, optionally, the covariances
(see the COV option) of the LS-means.
For more information, see the "Output Data Sets" section.
- PDIFF<=difftype>
-
requests that p-values for differences of the LS-means be
produced. The
optional difftype specifies which differences to display.
Possible values for difftype are ALL, CONTROL, CONTROLL, and CONTROLU. The ALL value
requests all pairwise differences, and it is the default. The CONTROL value
requests the differences with a control that, by default, is the first
level of each of the specified LS-mean effects.
To specify which levels of the effects are the controls, list
the quoted formatted values in parentheses after the keyword
CONTROL. For example, if the effects A, B, and C are
class variables, each having two levels, '1' and '2',
the following LSMEANS statement specifies the '1' '2' level
of A*B and the '2' '1' level of B*C as controls:
lsmeans A*B B*C / pdiff=control('1' '2', '2' '1');
For multiple effect situations such as this one, the ordering
of the list is significant, and you should check the
output to make sure that the controls are correct.
Two-tailed tests and confidence limits are associated with the CONTROL
difftype. For one-tailed results, use either the CONTROLL or
CONTROLU difftype. The CONTROLL difftype tests whether the noncontrol levels
are significantly less than the control; the lower confidence
limits for the control minus the noncontrol levels are considered to
be minus infinity. Conversely, the
CONTROLU difftype tests
whether the noncontrol levels are significantly greater than the control;
the upper confidence limits for the noncontrol levels minus the
control are considered to be infinity.
The default multiple comparisons adjustment for each difftype is
shown in the following table.
|
difftype
|
Default ADJUST=
|
| Not specified | T |
| ALL | TUKEY |
| CONTROL | |
| CONTROLL | DUNNETT |
| CONTROLU | |
If no difftype is specified, the default for the ADJUST= option is T
(that is, no adjustment); for PDIFF=ALL, ADJUST=TUKEY is the default;
in all other instances, the default value for the ADJUST= option is DUNNETT.
If there is a conflict between the PDIFF= and
ADJUST= options, the ADJUST= option takes precedence.
For example, in order to compute one-sided confidence limits for differences
with a control, adjusted according to Dunnett's procedure, the following
statements are equivalent:
lsmeans Treatment / pdiff=controll cl;
lsmeans Treatment / pdiff=controll cl adjust=dunnett;
- SLICE = fixed-effect
- SLICE = (fixed-effects)
-
specifies effects within which to test for differences between interaction
LS-mean effects. This can produce what are known as tests of simple
effects (Winer 1971). For example, suppose that A*B is significant and you
want to test for the effect of A within each level of B. The
appropriate LSMEANS statement is
lsmeans A*B / slice=B;
This code tests for the simple main effects of A for B, which are
calculated by extracting the appropriate rows from the coefficient
matrix for the A*B LS-means and using them to form an
F-test as performed by the CONTRAST statement.
- SINGULAR=number
-
tunes the estimability checking.
If ABS(L - LH) > C×number
for any row, then L is declared nonestimable.
H is the (X'X)-X'X
matrix, and C is ABS(L) except for rows where
L is zero, and then it is 1.
The default value for the SINGULAR= option is 10-4.
Values for the SINGULAR= option must be between 0 and 1.
- STDERR
-
produces the standard error of the LS-means and the
probability level for the hypothesis H0: LS-mean = 0.
- TDIFF
-
produces the t values for all hypotheses
H0: LS-mean(i) = LS-mean(j) and the corresponding probabilities.
Copyright © 1999 by SAS Institute Inc., Cary, NC, USA. All rights reserved.
