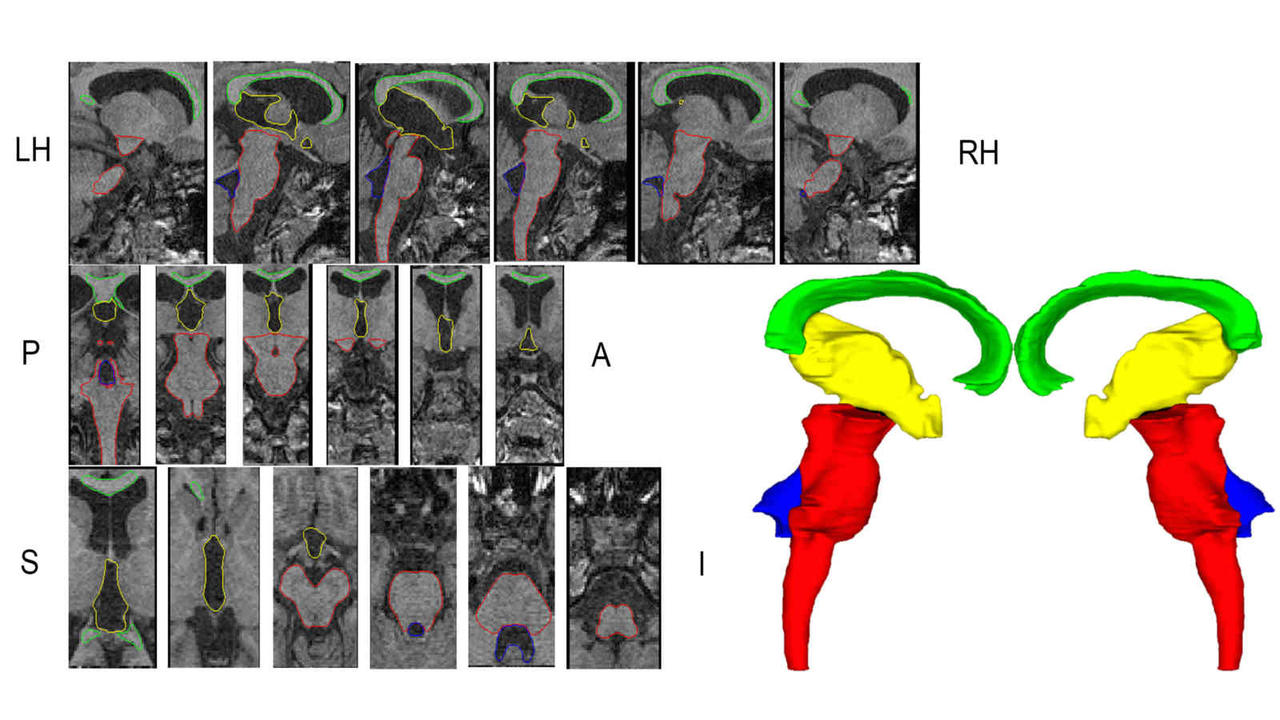
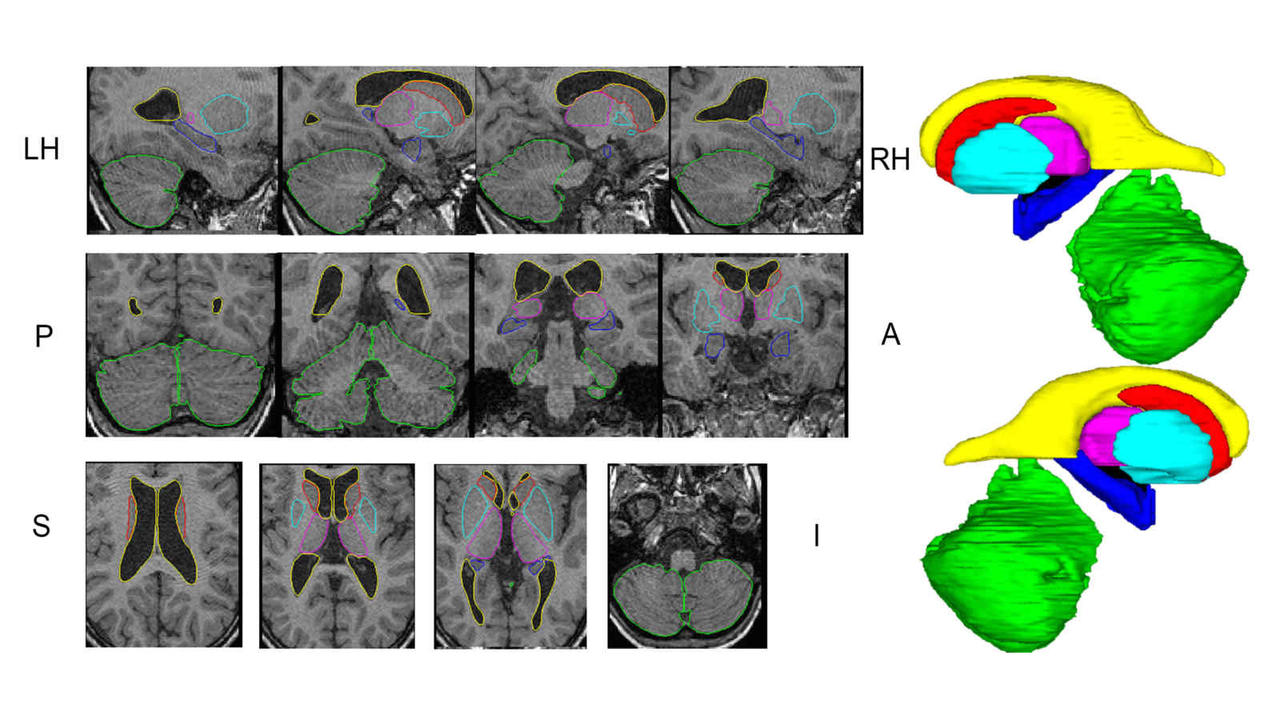
Research focus
- Computational Brain Anatomy
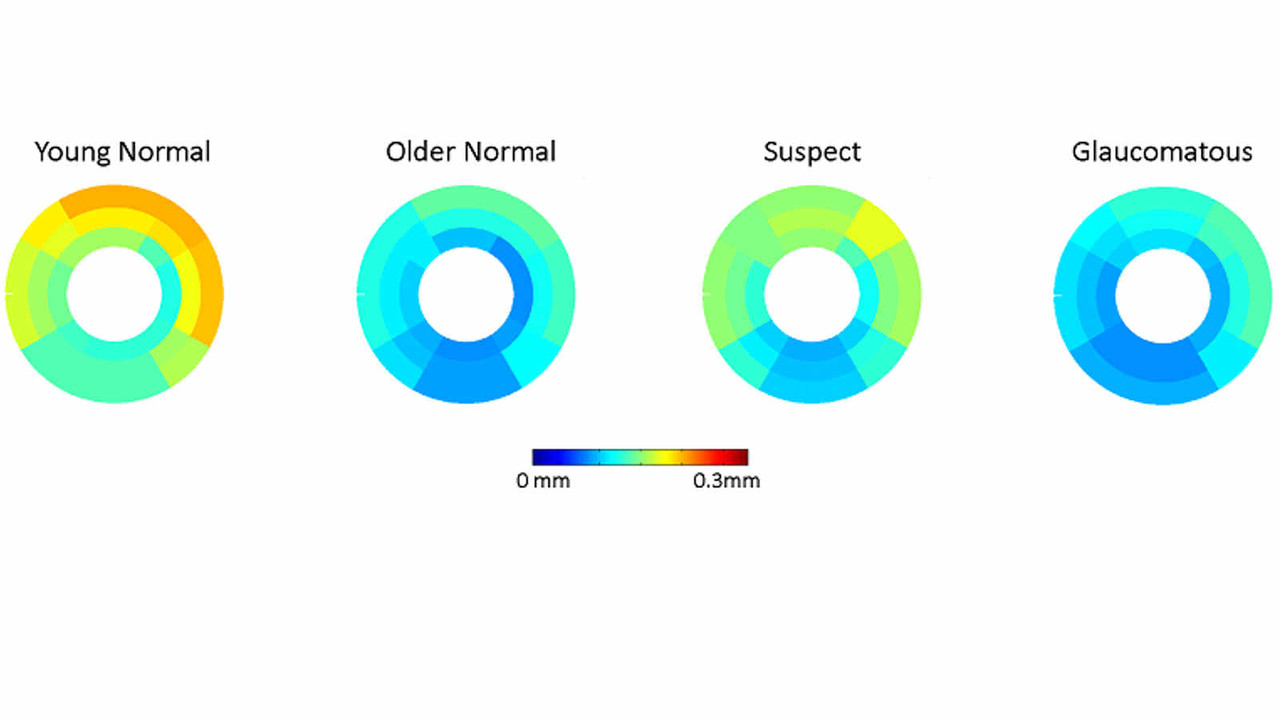
- Computational Eye Anatomy
- Image Registration and Segmentation
- Image Acquisition
- Quality Check Visualizations
- Sponsors & Funding
Computational Brain Anatomy
Network Analysis
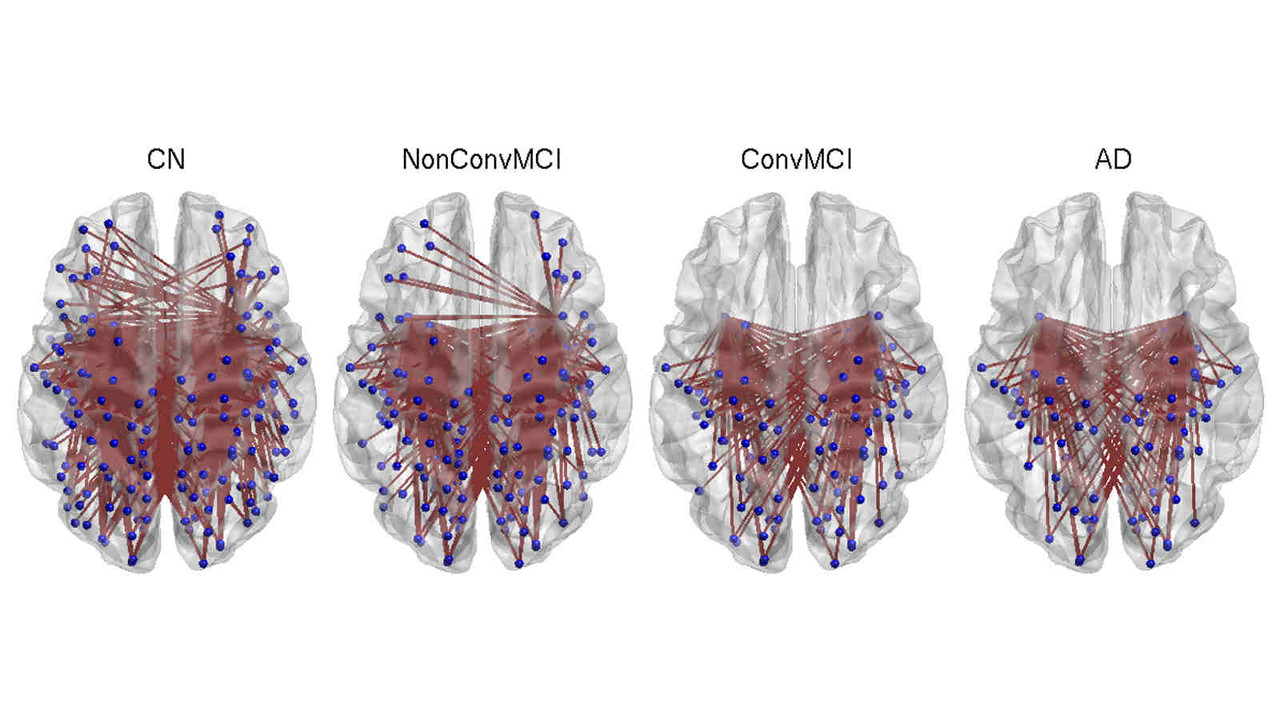
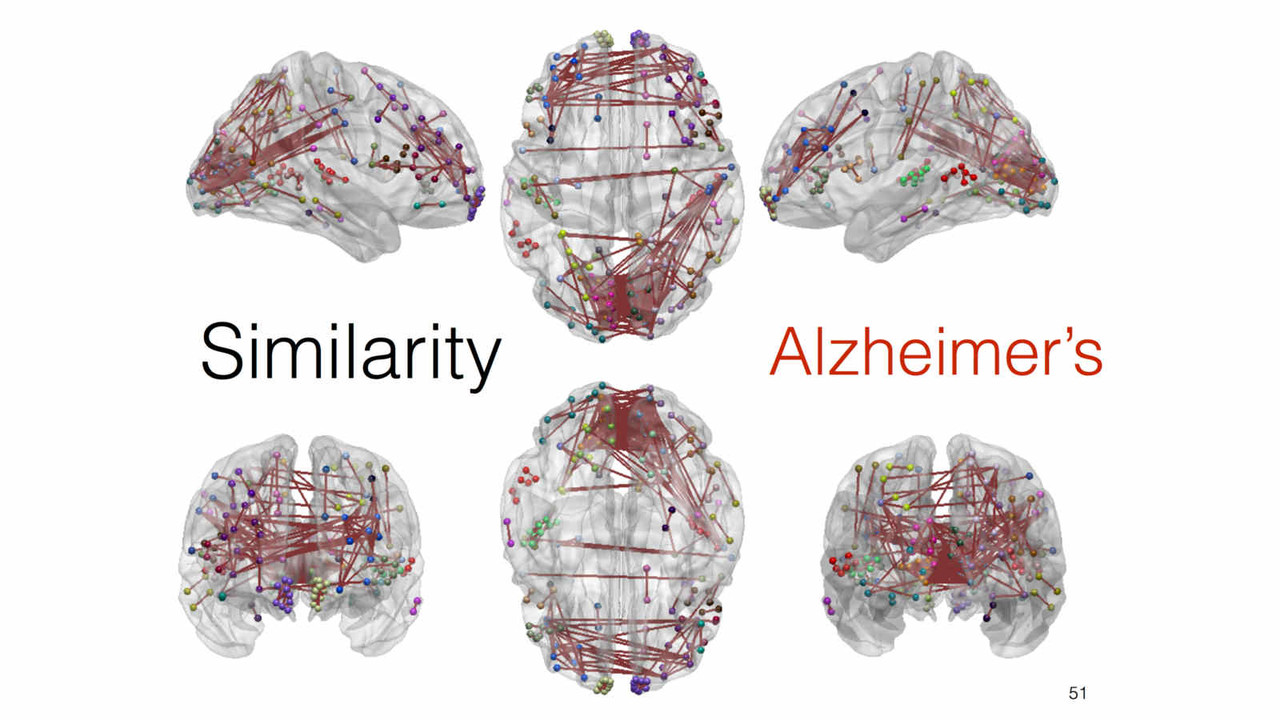
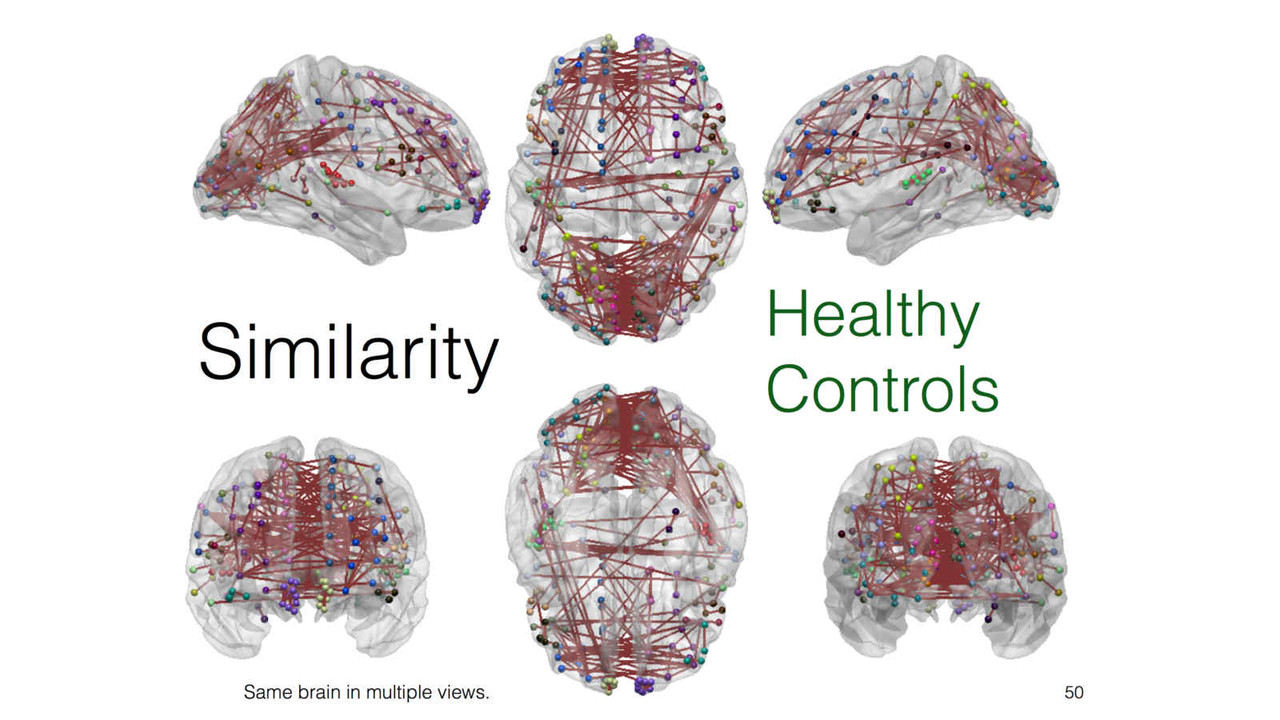
In this stream of research, we explore and investigate novel methods of extracting new insights into a given feature such as cortical thickness, via network-level analysis. Analyzing the inter-regional covariance in a given feature povides with us a richer description of the underlying patterns, and network-level changes caused by neurodegenerative diseases such as Alzheimer's disease.
Related publicationsImaging Genetics
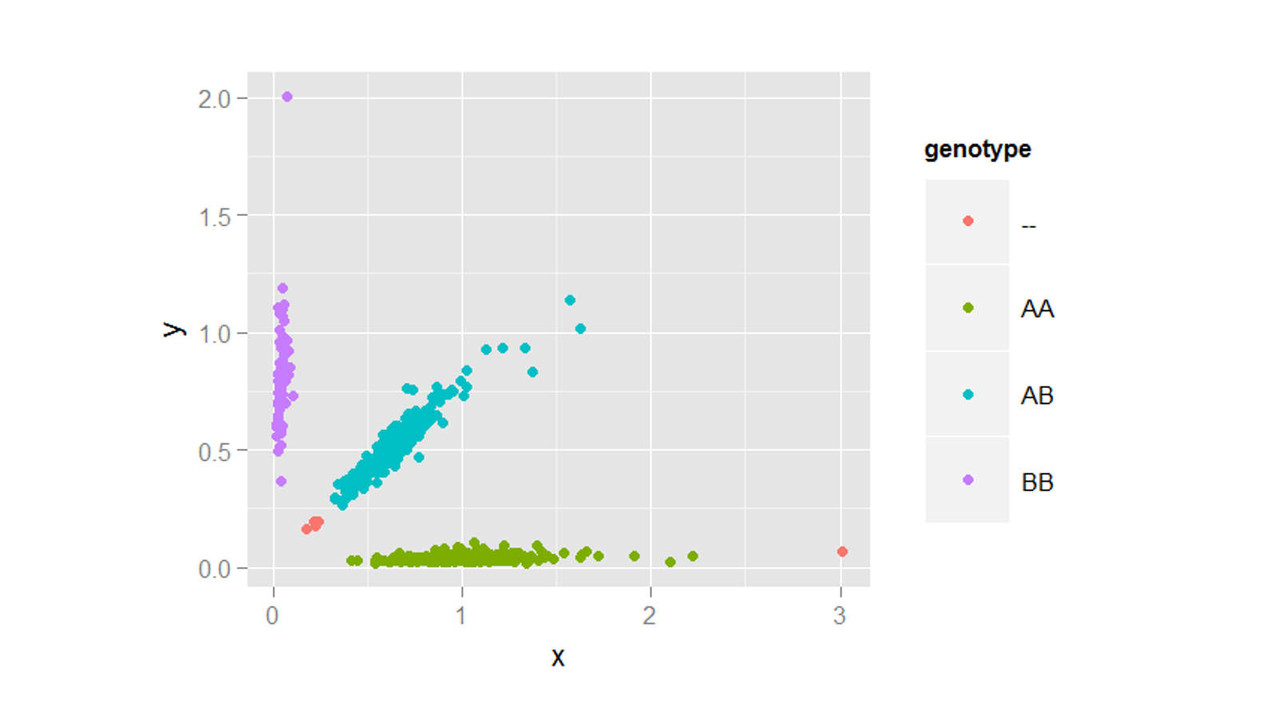
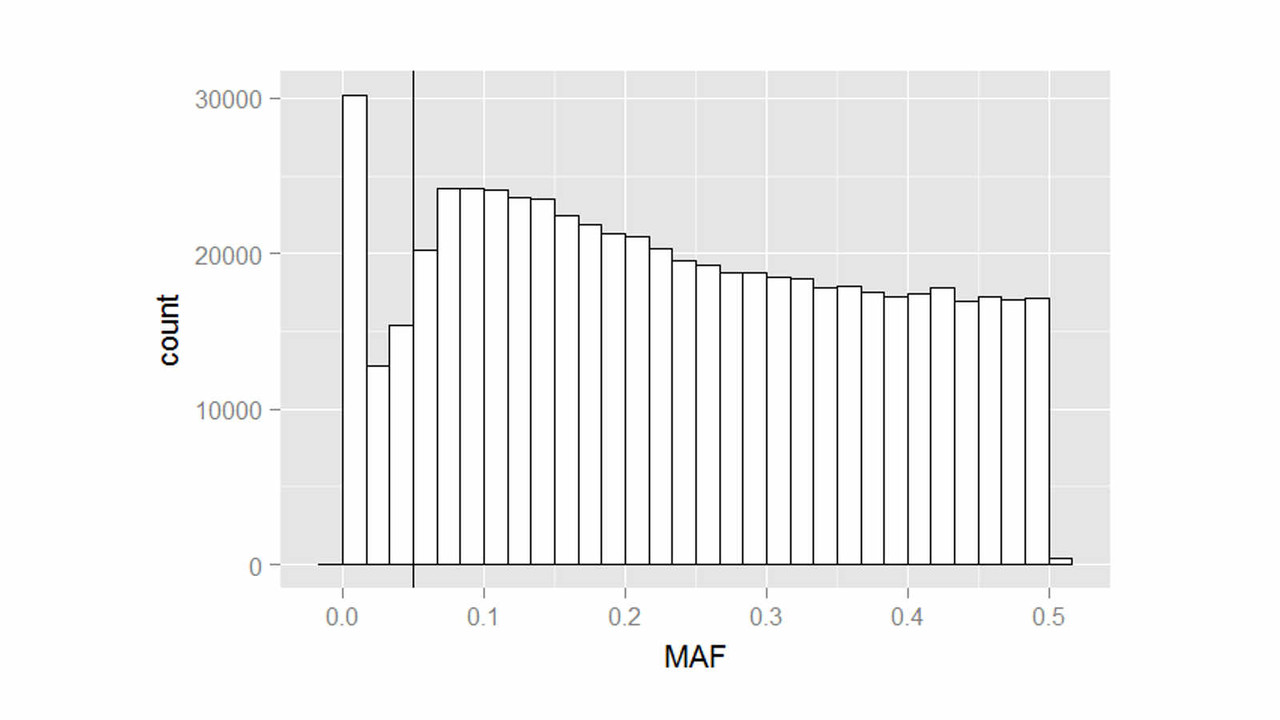
Both genetic variants and brain region abnormalities are recognized to play a role in cognitive decline. In this project, we explore the relationship between genome-wide variation and region-specific rates of decline in brain structure, as measured by magnetic resonance imaging.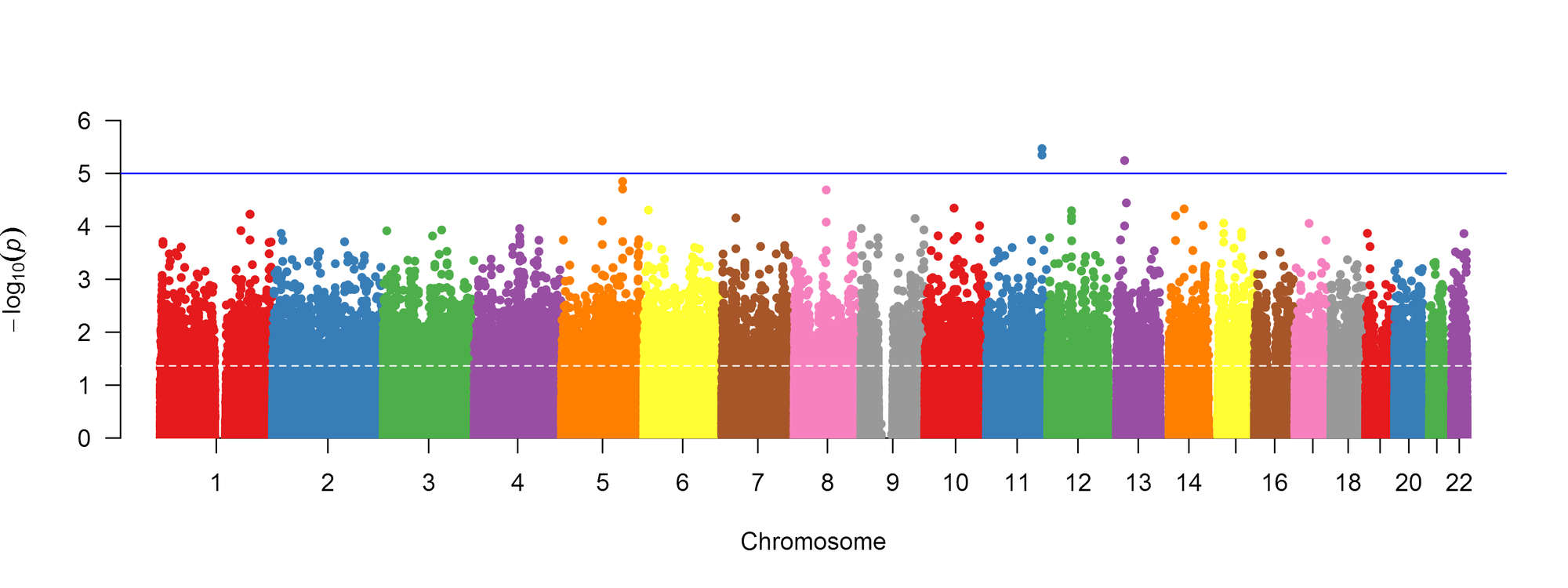
Above: Manhattan plot of association between genetic markers and cognitive impairment, adjusted for confounders.
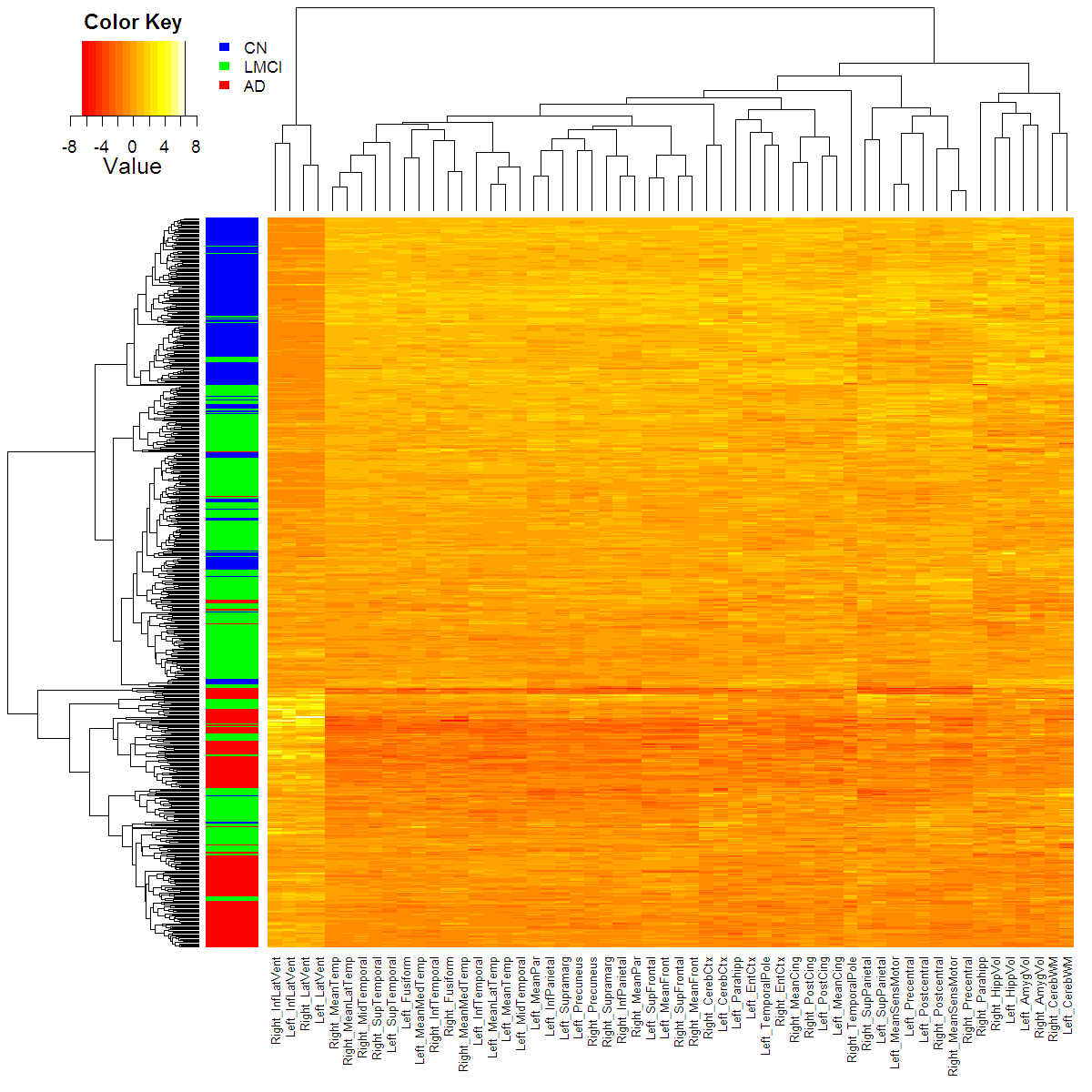
Below: Heatmap of the rates of change in MRI features with coloured disease labels, adjusted for confounders.
Alzeheimer's Disease
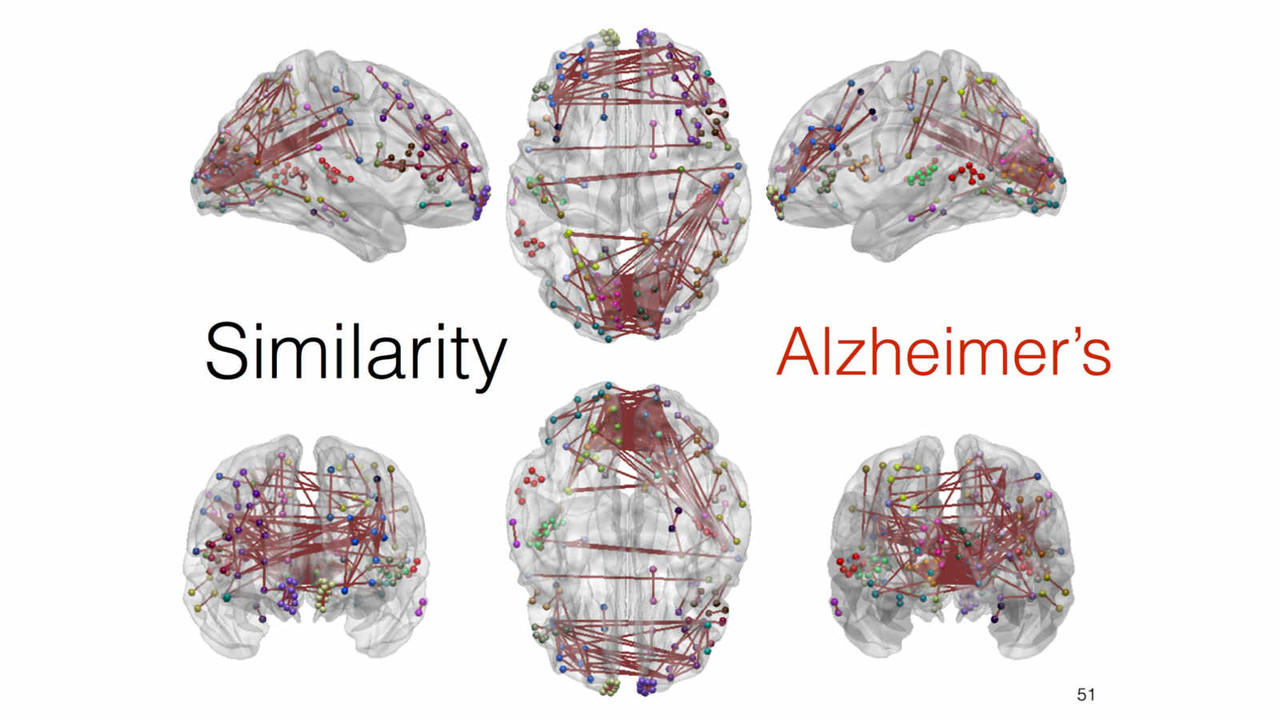
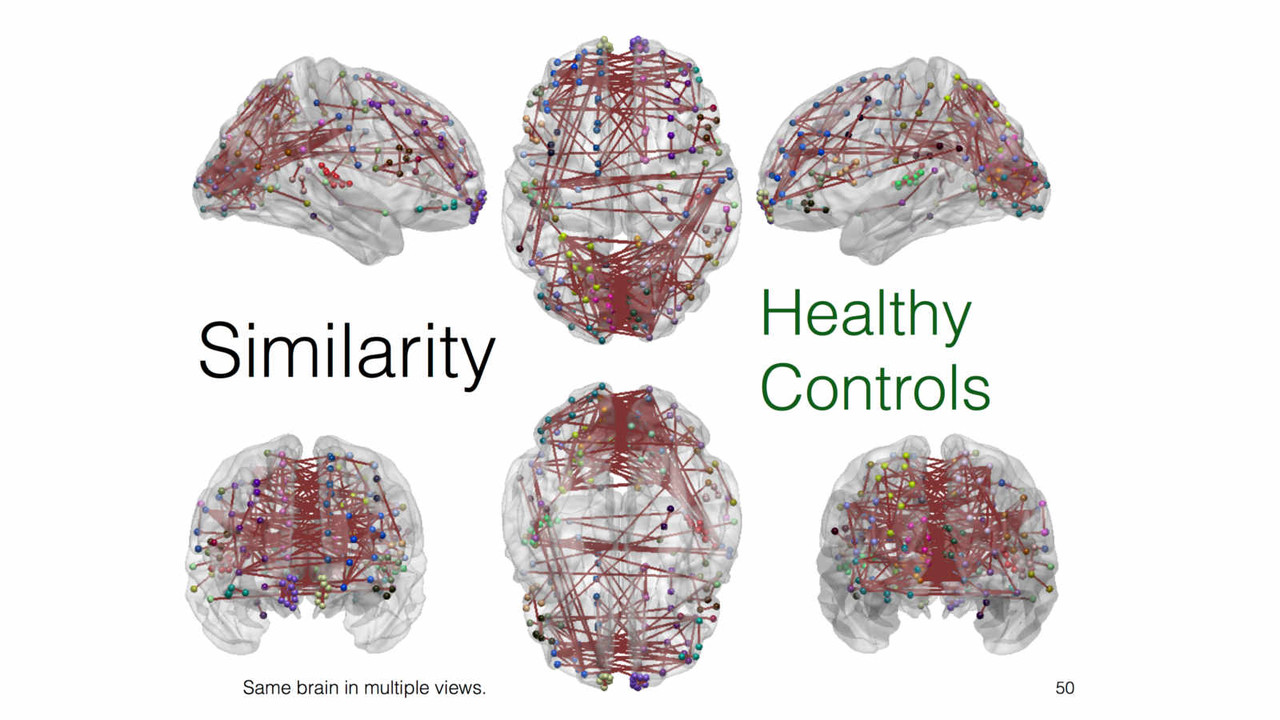
A key focus of our laboratory is on the development of fully automated computational-anatomy tools for the early detection of Alzheimer's disease. Towards this challenging goal, we develop various method through the spectrum of medical image analysis starting from organ segmentation, accurate registration, feature extraction methods and machine learning tools. The above picture shows a comparison of novel T1-MRI based networks we have constructed to help build new covariance features which show promise for the early detection of Alzheimer's disease. Please visit pages in the Research section to learn more about our research.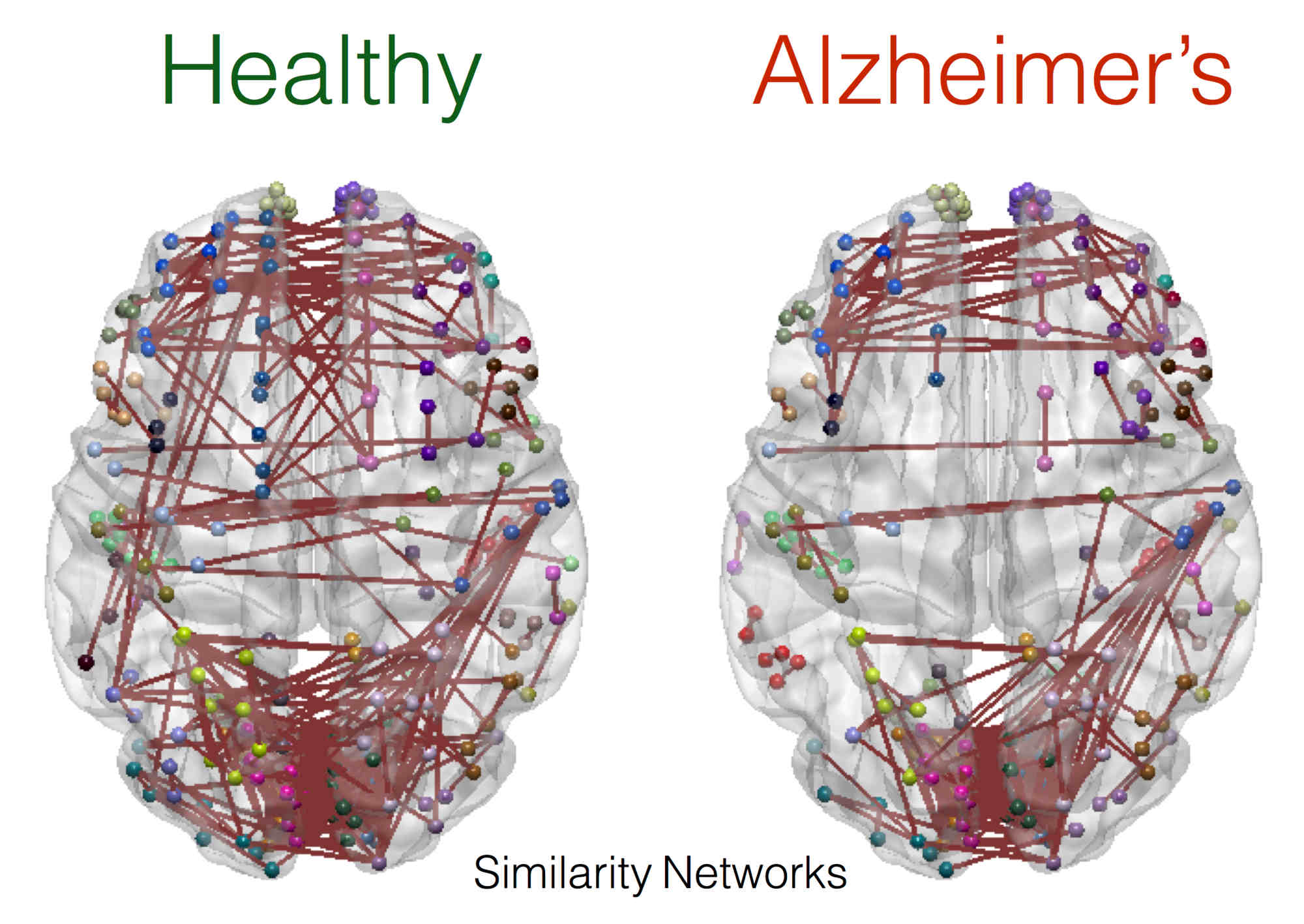
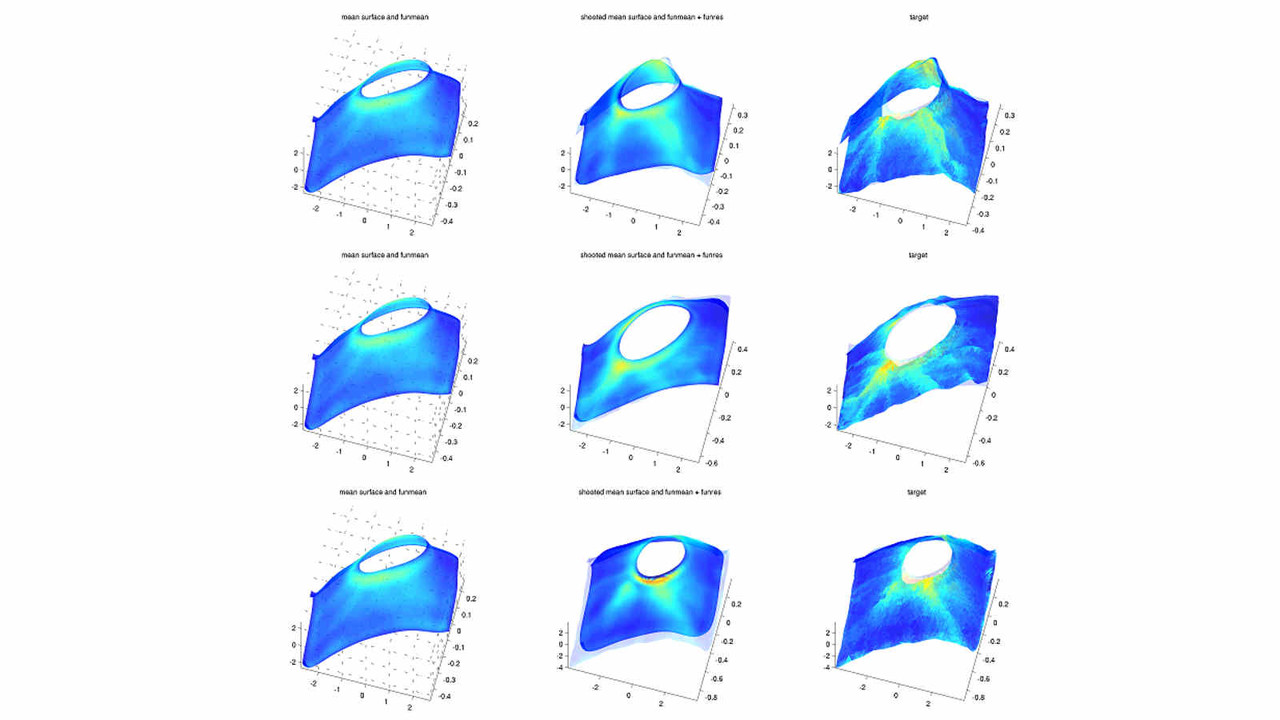
Functional Shape Analysis
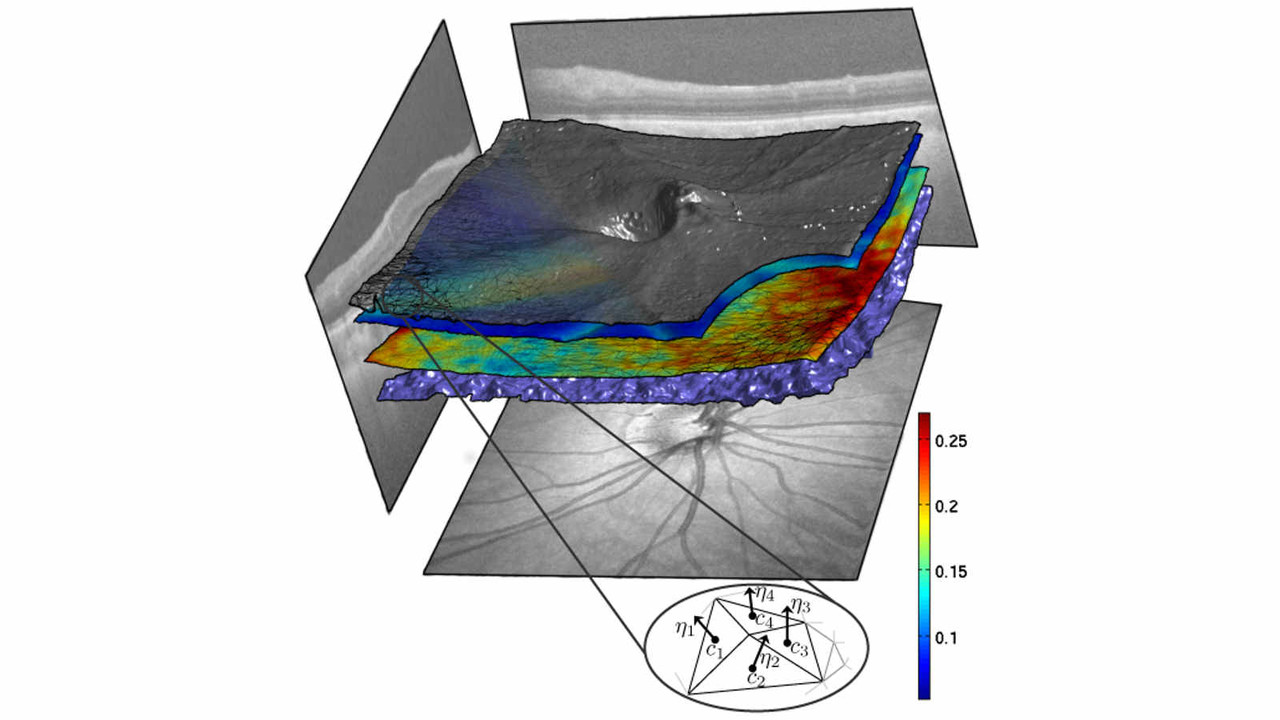
 In this project, we employ the novel functional shape analysis (fshape) paradigm for the characterization of shape variability in cortical and subcortical regions extracted from MRI images in the brain, among healthy individuals, and patients suffering with Alzheimer's disease (AD) and related dementias.
In this project, we employ the novel functional shape analysis (fshape) paradigm for the characterization of shape variability in cortical and subcortical regions extracted from MRI images in the brain, among healthy individuals, and patients suffering with Alzheimer's disease (AD) and related dementias.
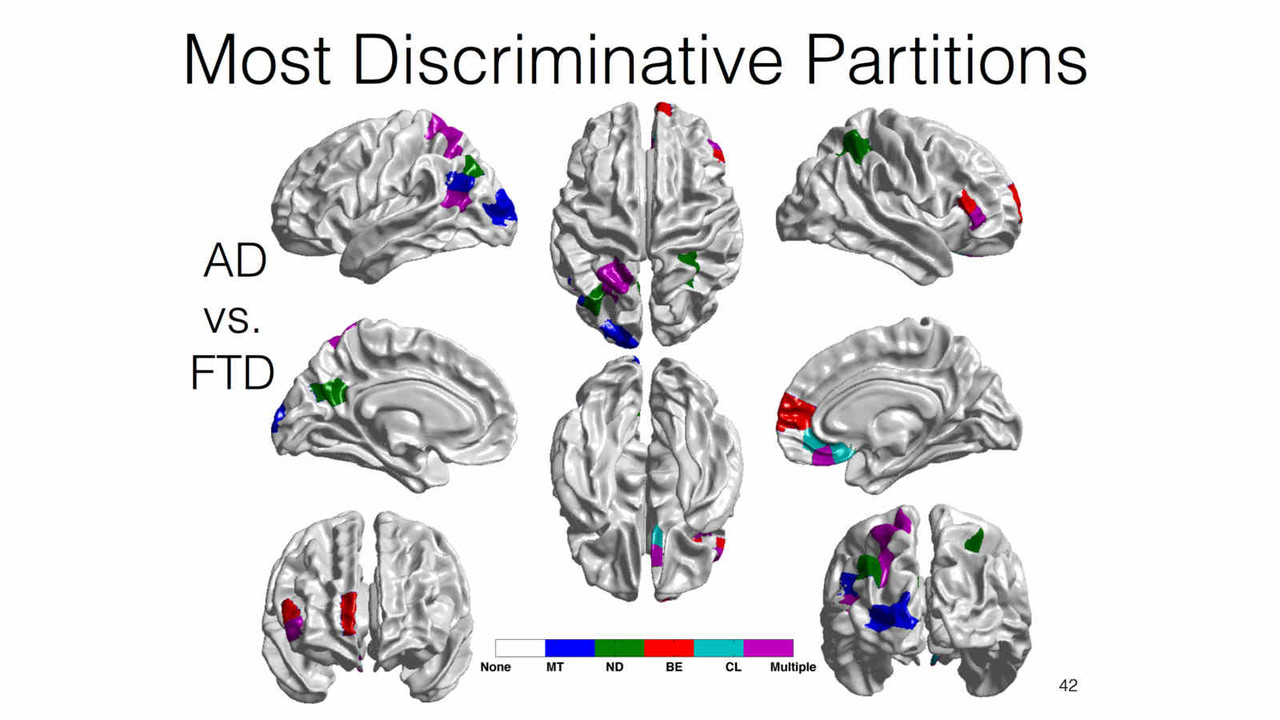
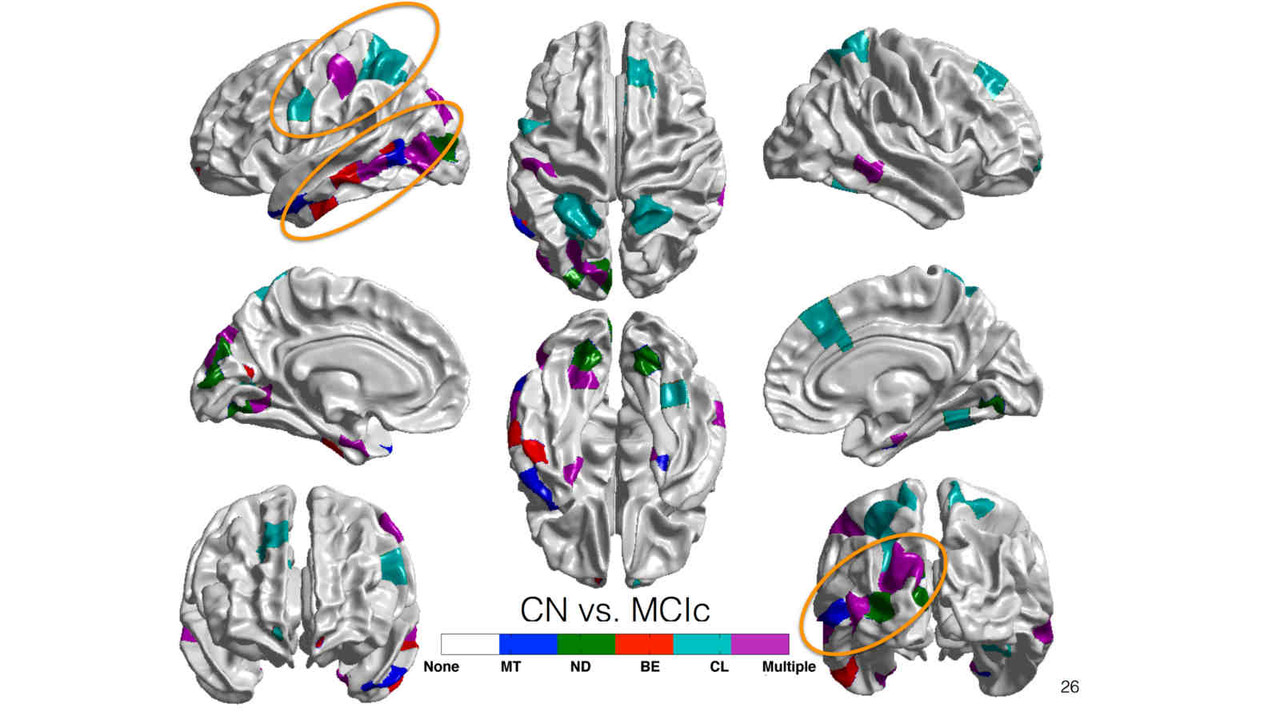
Differential Diagnosis
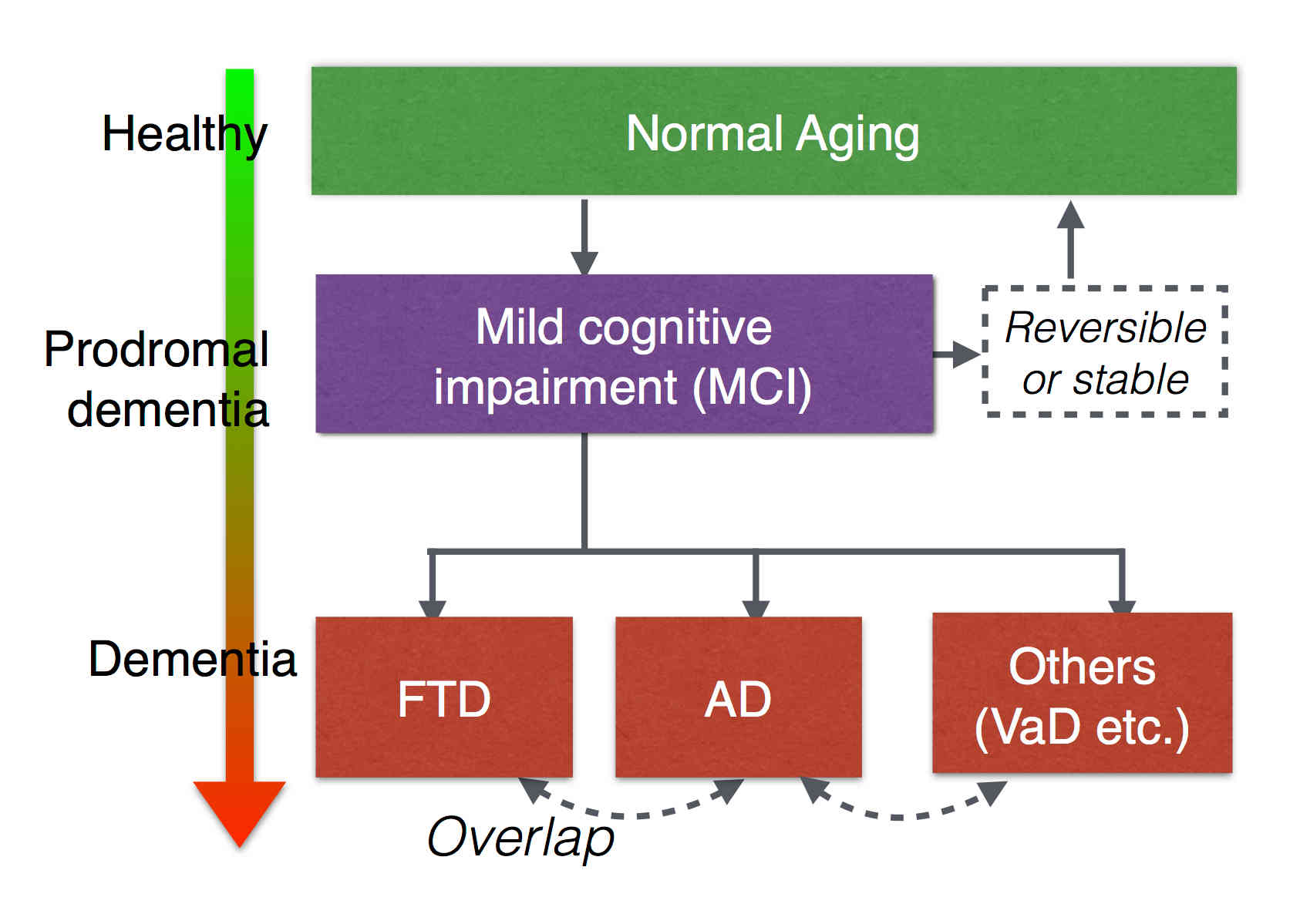
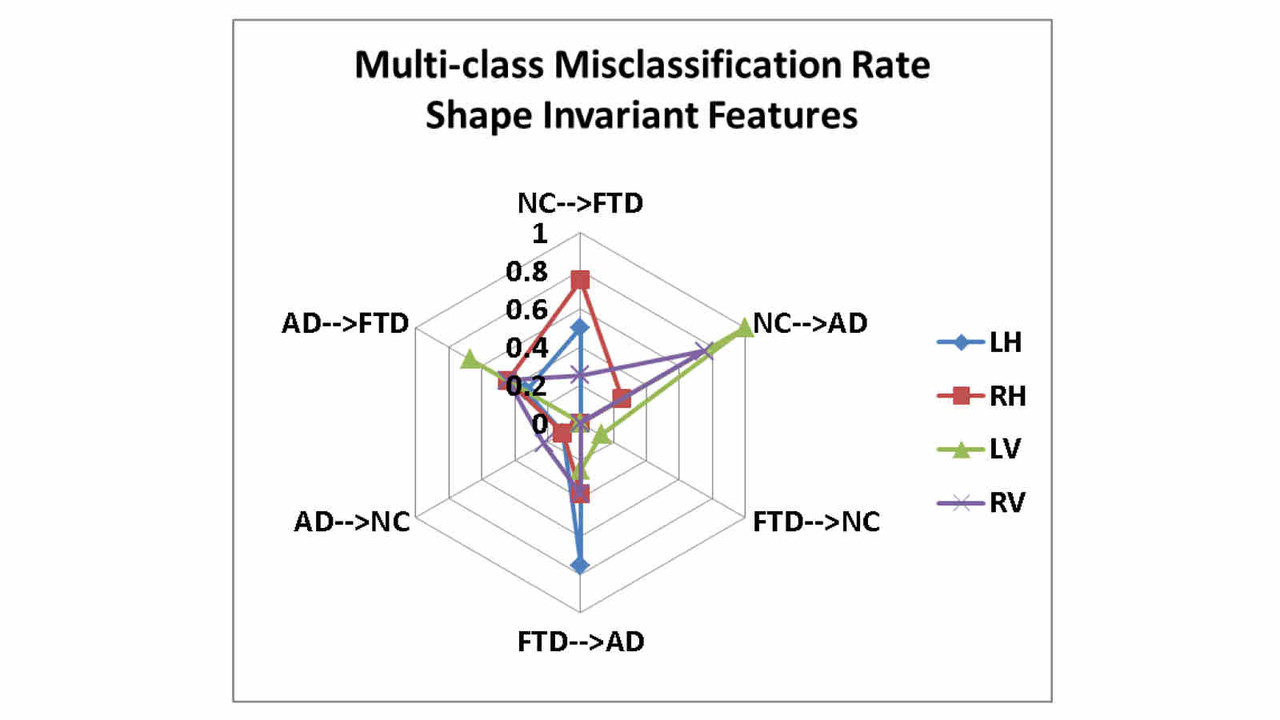
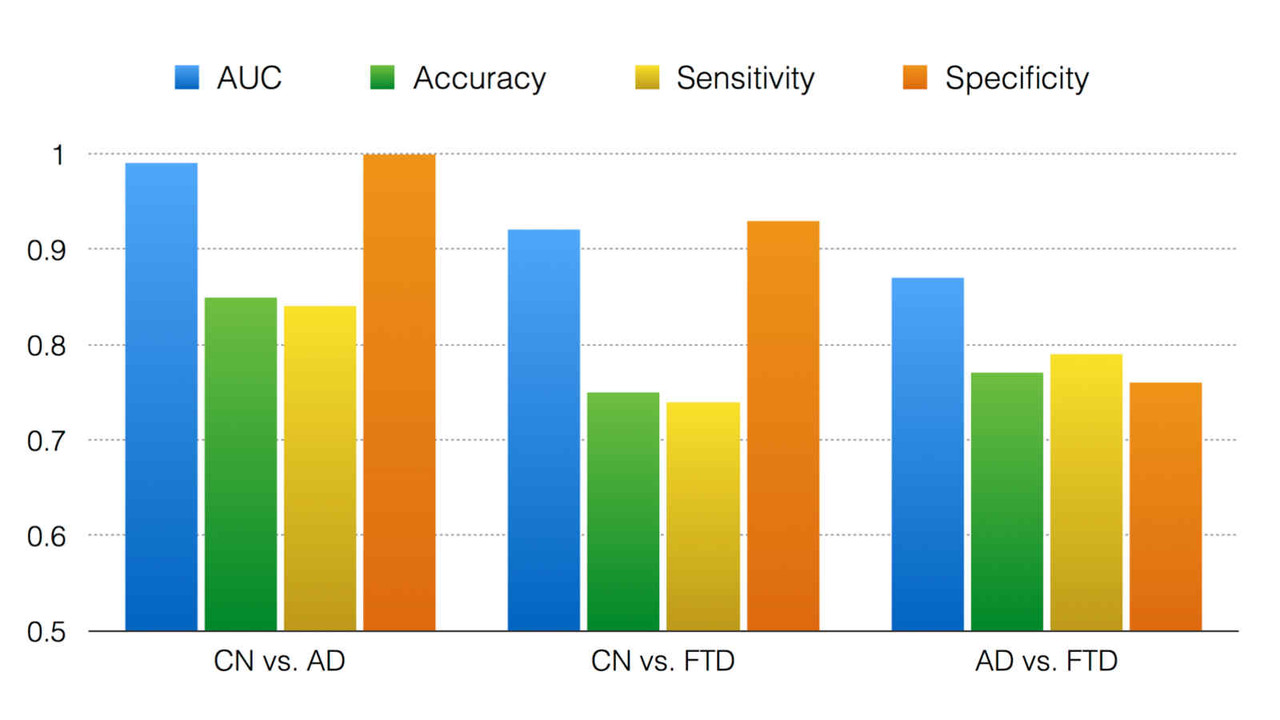
Biomarkers derived from brain magnetic resonance (MR) imaging have promise in being able to assist in the clinical diagnosis of brain pathologies. These have been used in many studies in which the goal has been to distinguish between pathologies such as Alzheimer’s disease and healthy aging. However, other dementias, in particular, frontotemporal dementia, also present overlapping pathological brain morphometry patterns. In this stream of research, we investigate novel methods for the differential diagnosis of various neurodegenrative diseases in a multi-class setting, and explore novel methods and techniques to improve the differential diagnostic accuracy.
Related Publications:Pradeep Reddy, Howard Rosen, Bruce Miller, Michael Weiner, Lei Wang, Mirza Faisal Beg, Three class differential diagnosis among AD, FTD and controls, Frontiers in Neurol- ogy, 5(71), 2014.
Pediatric Shape Analysis
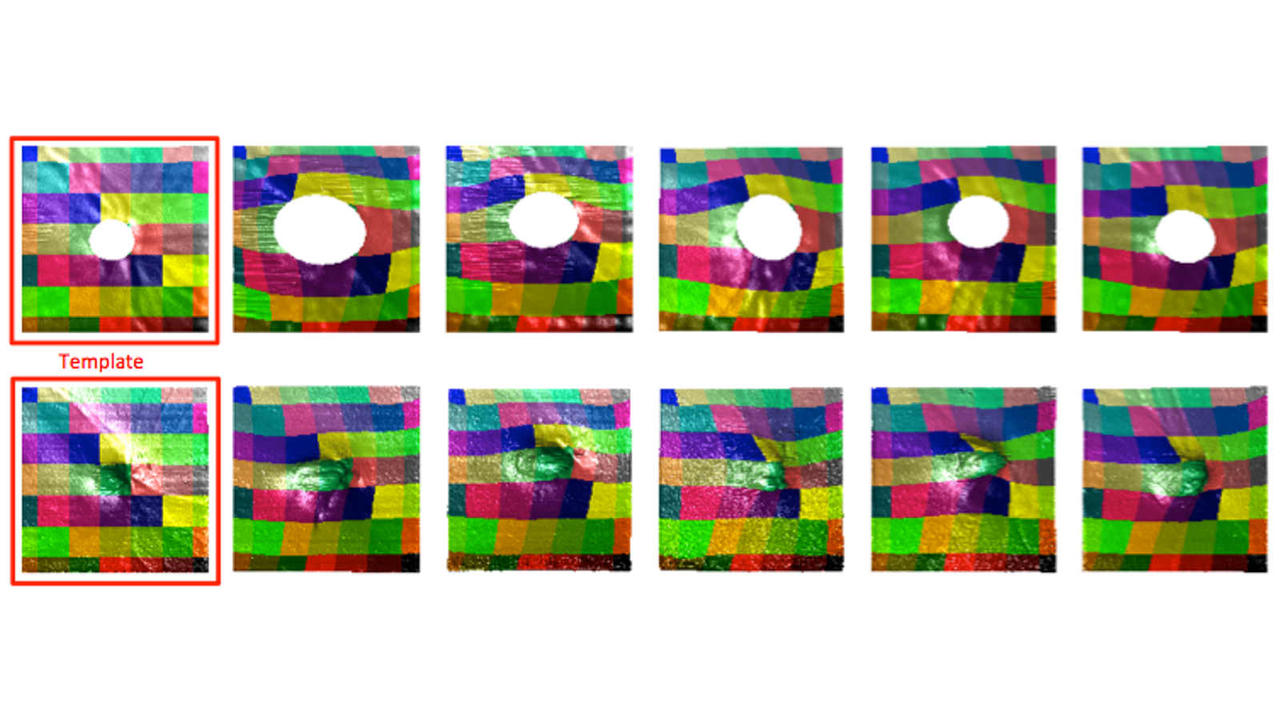
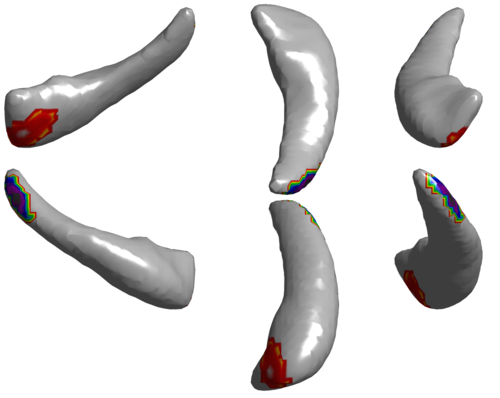 The change in shape of the anatomical structures within the brain is expected to have a profound effect on behavioral, learning and psychological outcomes. Brain development during early childhood from birth to adolescence is accompanied with increase in brain volume, neuronal growth in the cortex and shape formation of structures within the brain. In this work we aim to develop an automated method for analysis of brain MRI data and objective quantification of the change in shape within the brain with normal growth and the associated alterations due to diseased conditions.
The change in shape of the anatomical structures within the brain is expected to have a profound effect on behavioral, learning and psychological outcomes. Brain development during early childhood from birth to adolescence is accompanied with increase in brain volume, neuronal growth in the cortex and shape formation of structures within the brain. In this work we aim to develop an automated method for analysis of brain MRI data and objective quantification of the change in shape within the brain with normal growth and the associated alterations due to diseased conditions.
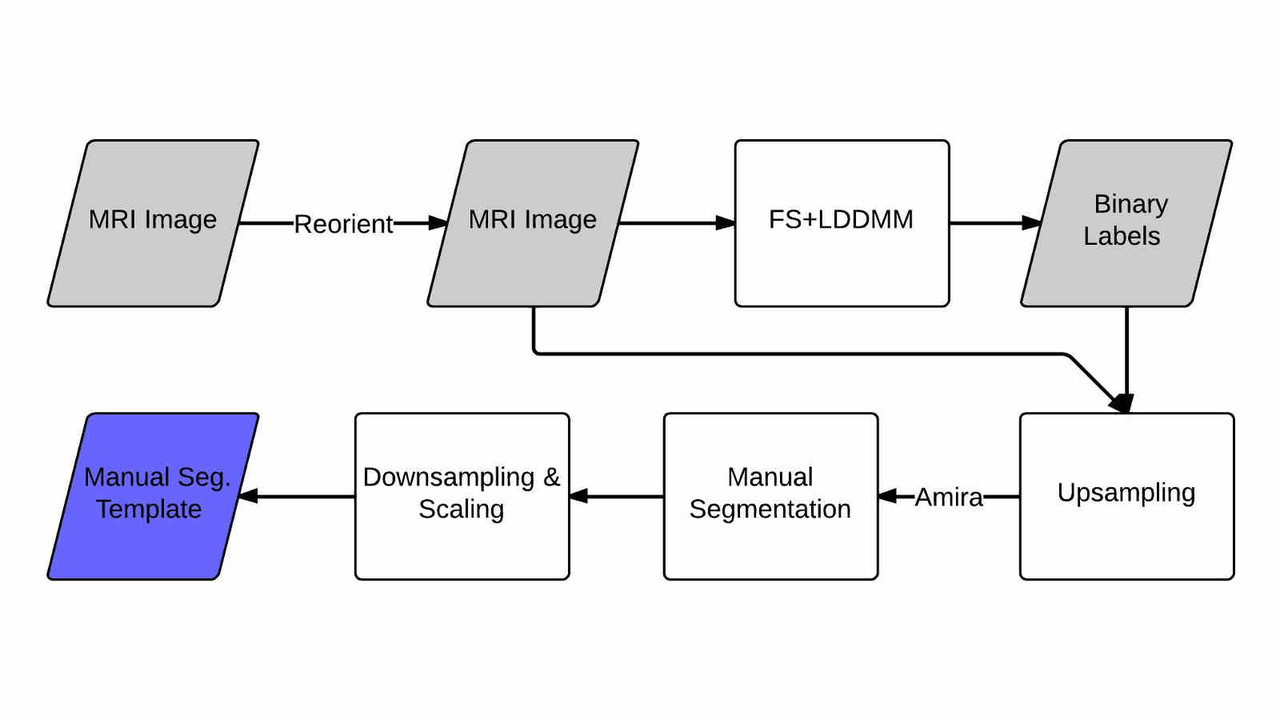
Pediatric Template Library
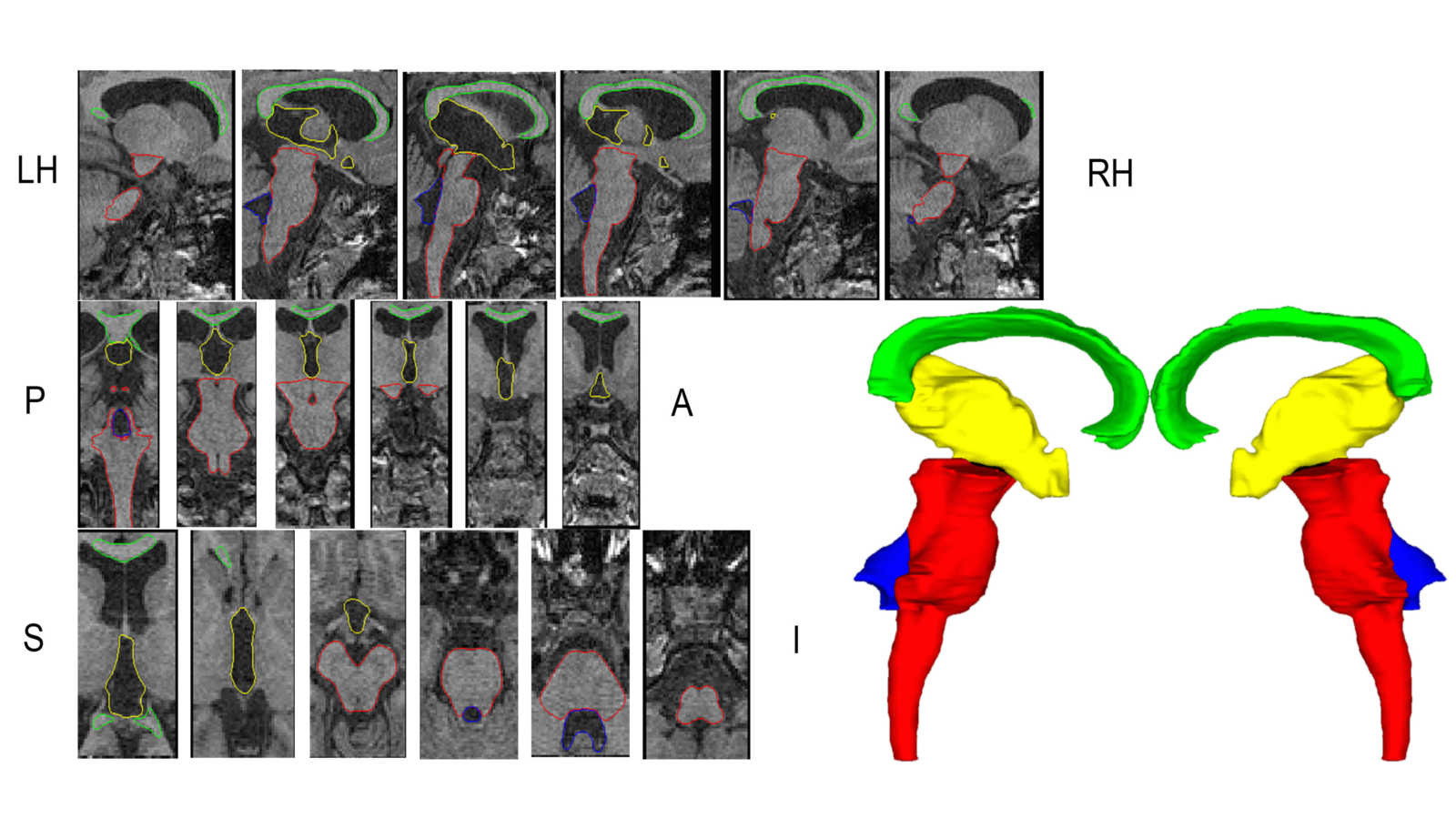
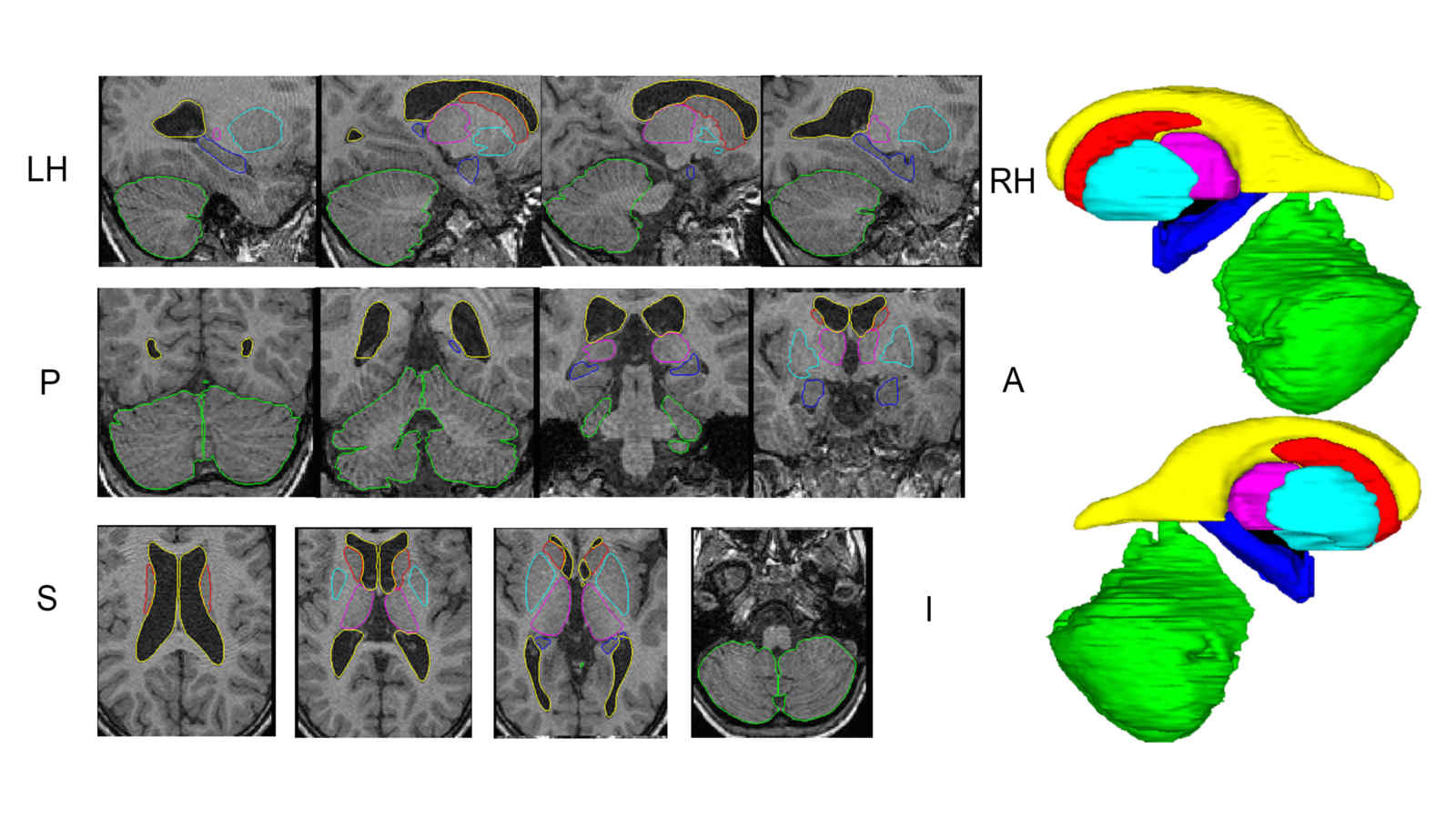
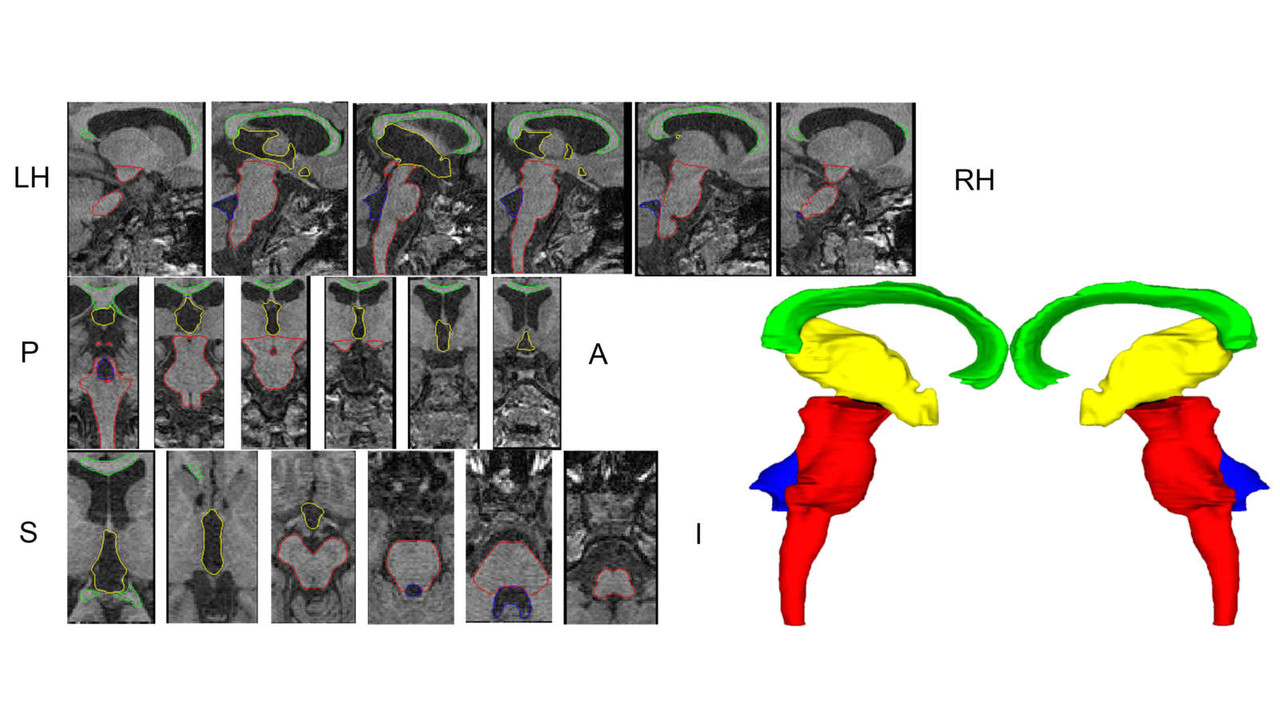
In this work we created a library of ground truth template data with manual segmentation labels for 16 subcortical structures in brain MRI data from children at 8-years of age.Publication: Manually segmented template library for 8-year-old pediatric brain MRI data with 16 subcortical structures
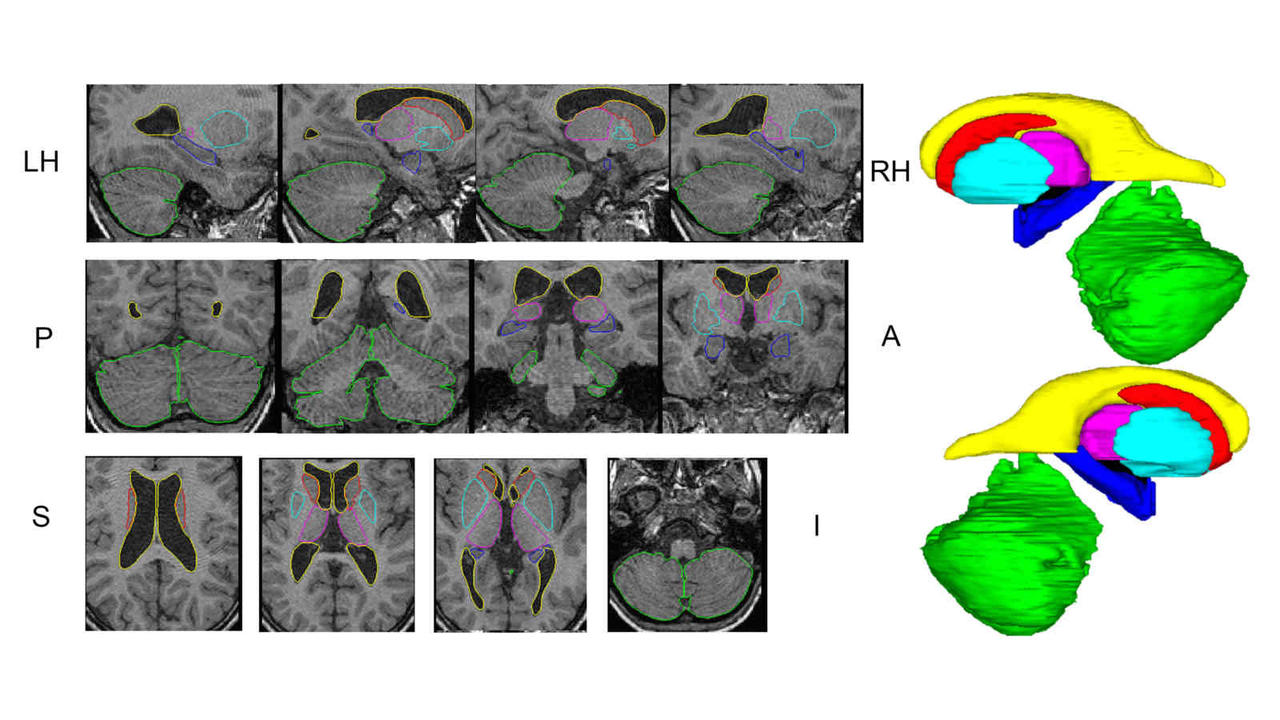
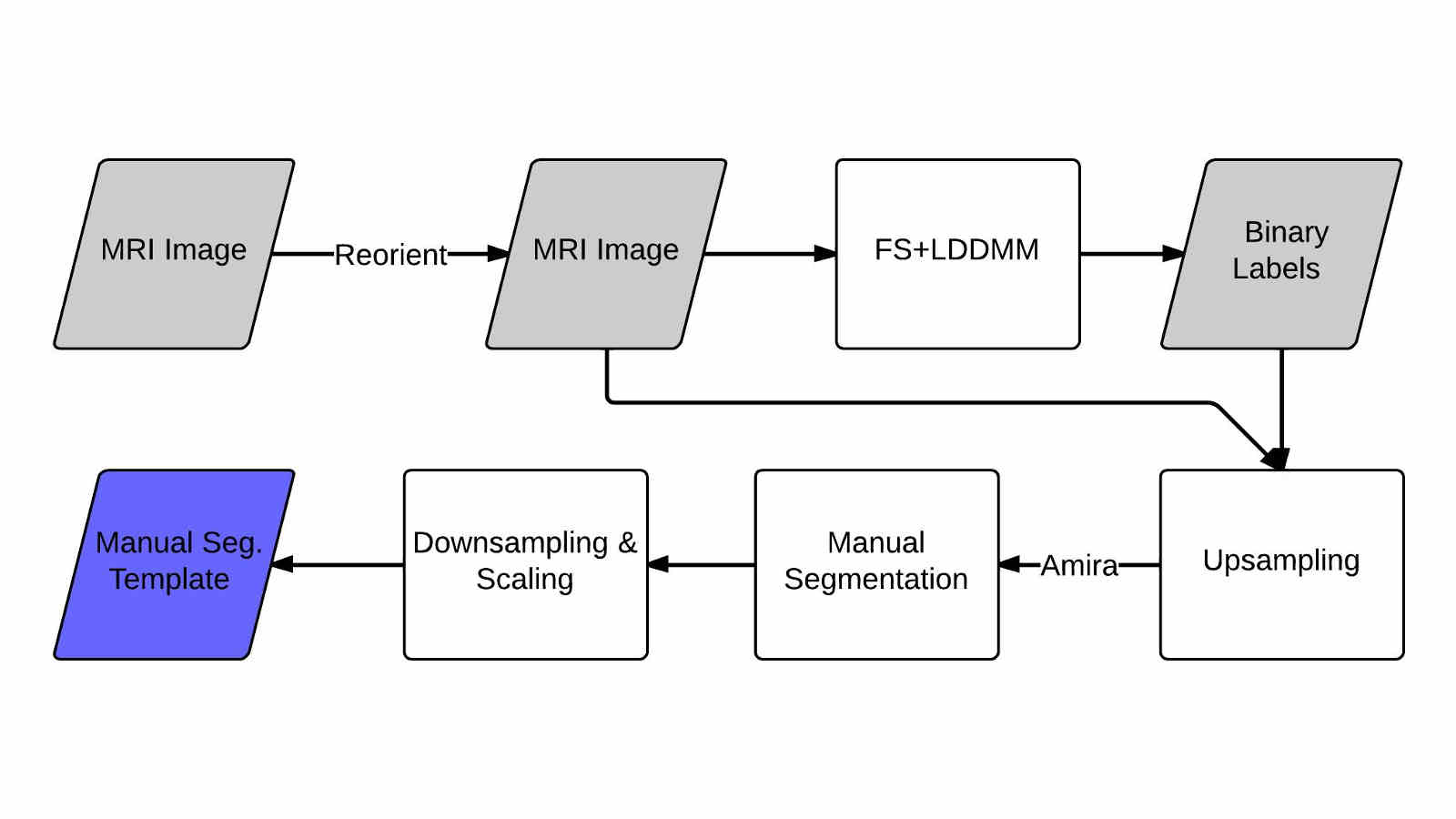 Above: The manual segmentation procedure followed for creation of the library of templates.
Above: The manual segmentation procedure followed for creation of the library of templates.
Below: The overlay of outlines of the structures on the respective MRI slices for an example subject.
Dimensionality Reduction of Cortical Thickness Measurements
Neural atrophy patterns in the cerebral cortex are closely correlated to noticeable cognition decline due to Alzheimer's disease. These patters of thinning can be analyzed by processing structural magnetic resonance images. Freesurfer software is used to automatically segment white and gray matter in the brain, label the cortical and subcortical regions, register all of the brains to a common template, perform cortical thickness parcellation and thickness map smoothing. Approximately 300,000 cortical thickness measurements are computed from the whole cortex of every patient. This data has dimension equal to the number of thickness measurements taken on each patient. In order to visualize this high-dimensional data and find features related to Alzheimer's disease we must reduce the dimensionality. We present a method for dimensionality reduction, consisting of anatomically subdividing the brain into patitions and performing principal component analysis to identify a small subset of the original variables that contain the most information about the variance in the data.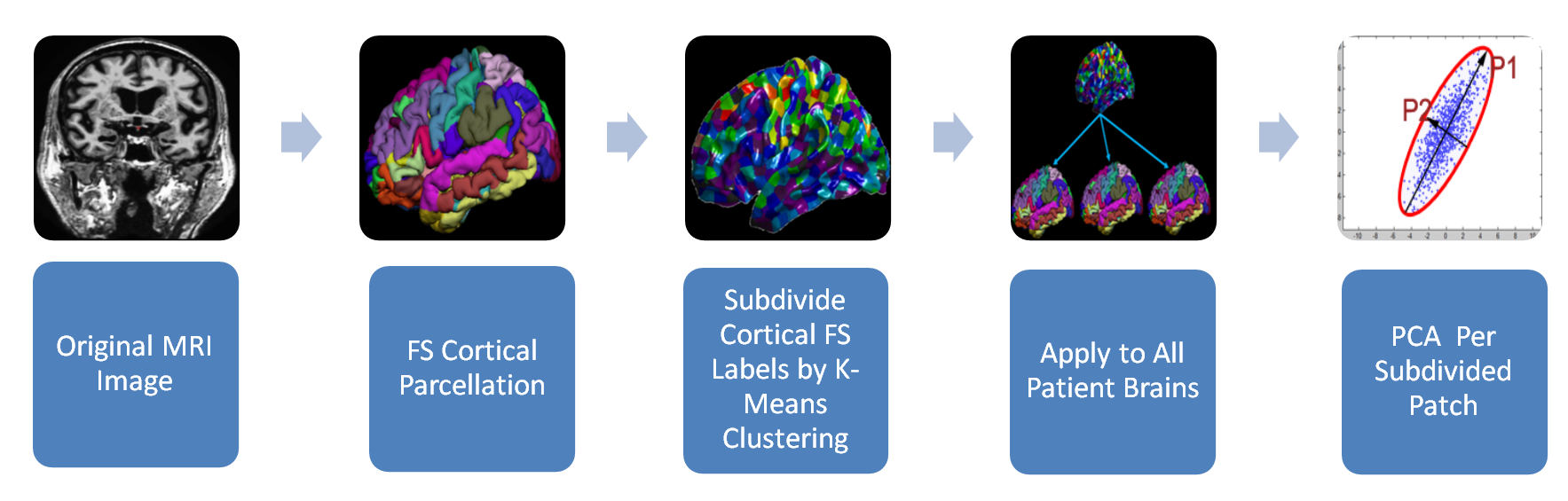 Figure 1. Pipeline of Dimensionality Reduction of Cortical Thickness Measurements Using Principal Component Analysis (PCA) of Patches of the Brain Generated by K-Means Clustering of Freesurfer (FS) Labels
Figure 1. Pipeline of Dimensionality Reduction of Cortical Thickness Measurements Using Principal Component Analysis (PCA) of Patches of the Brain Generated by K-Means Clustering of Freesurfer (FS) Labels
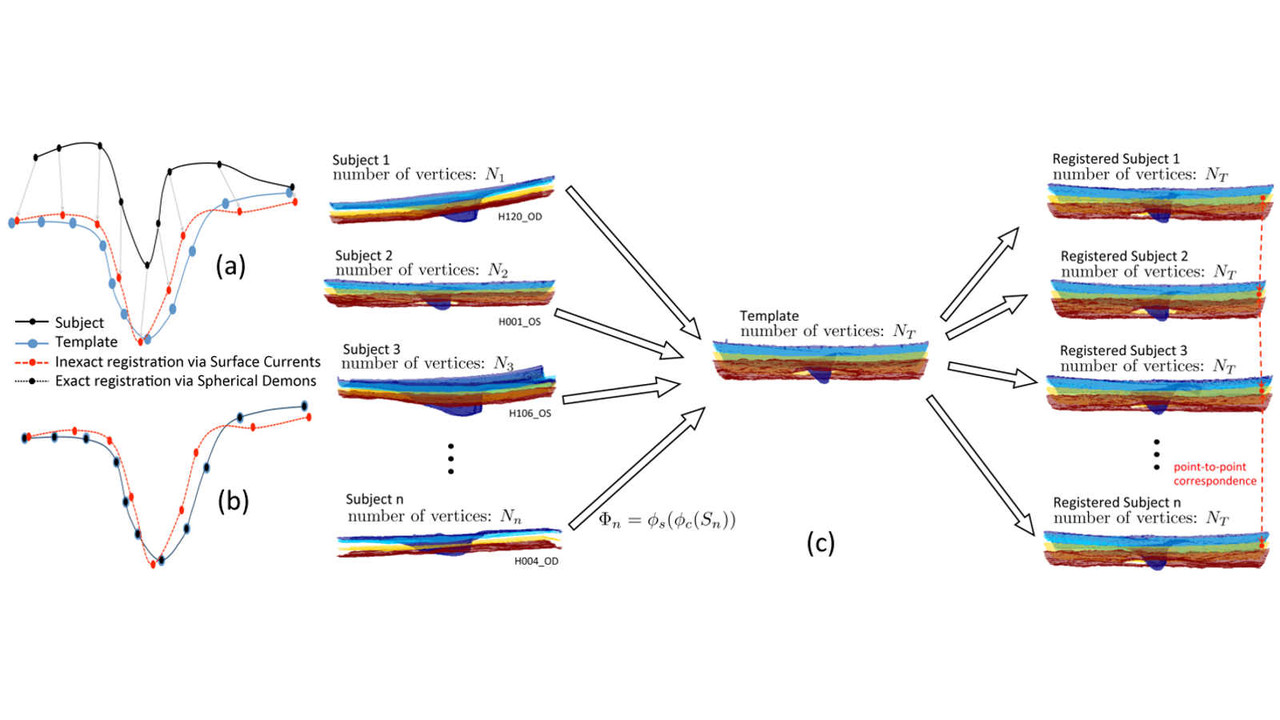
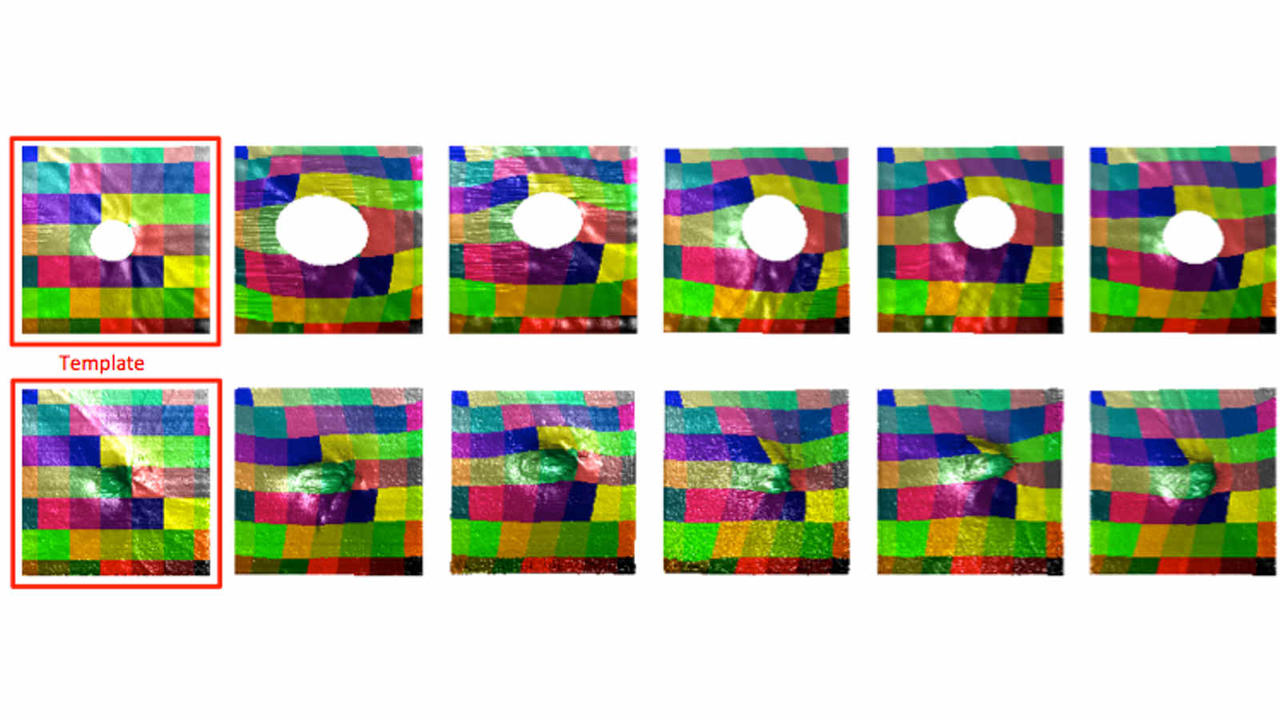
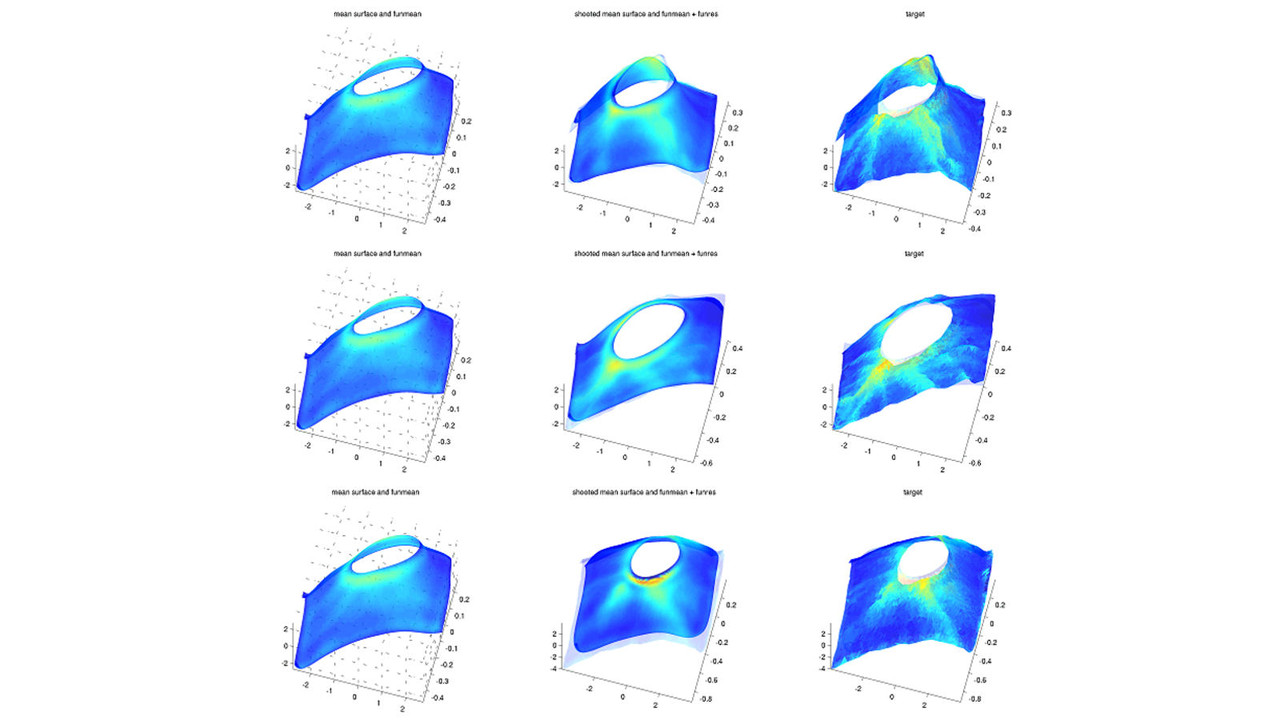
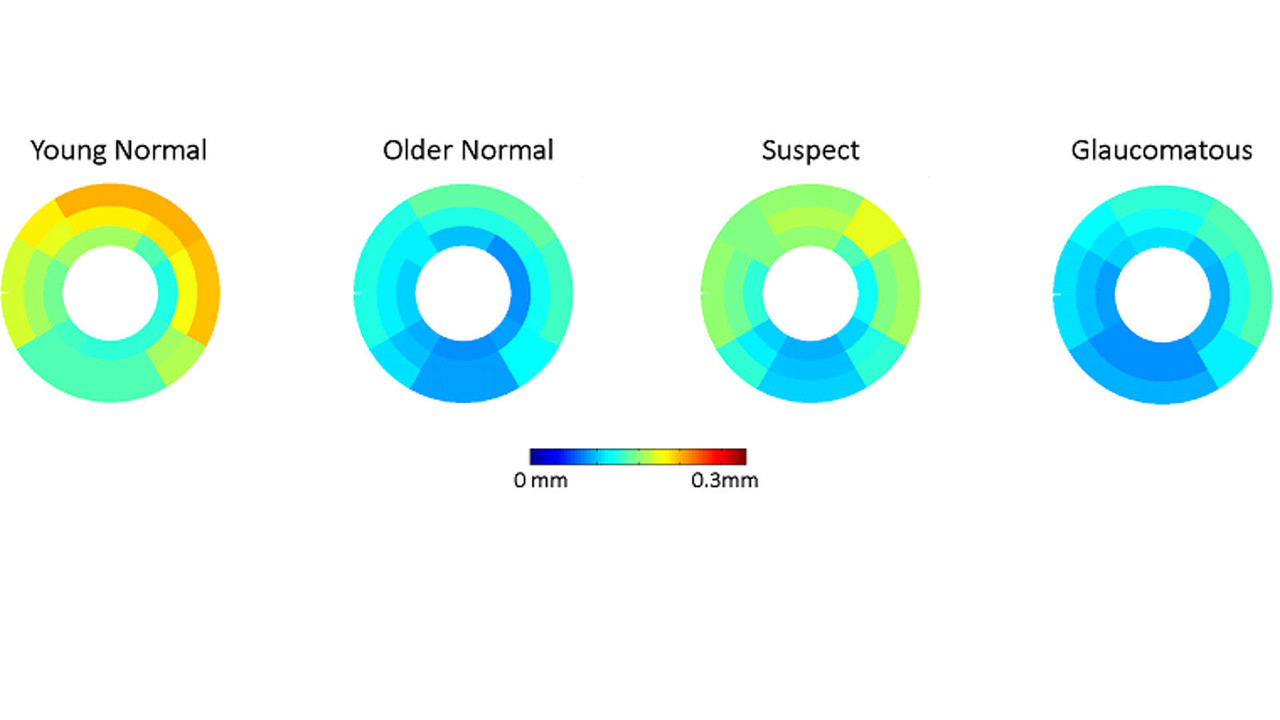
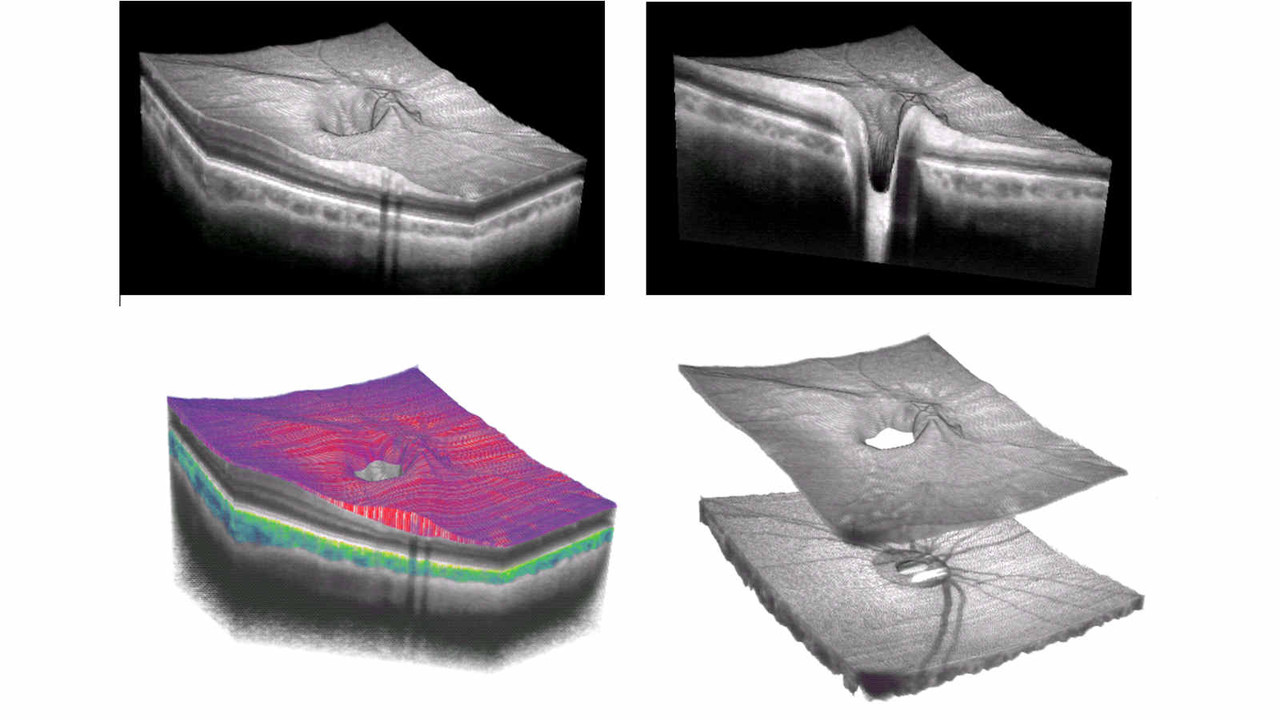
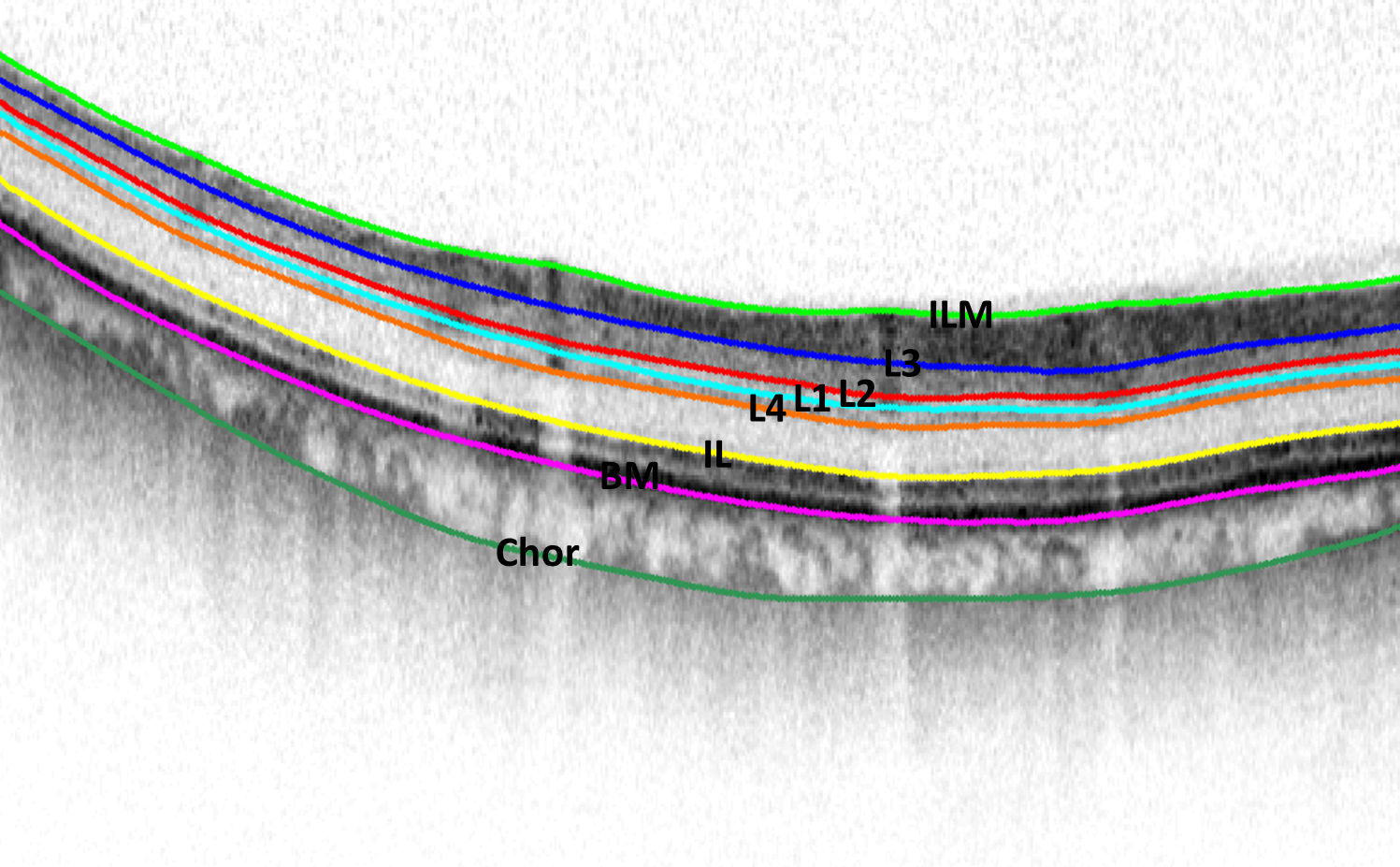 Publications
Publications
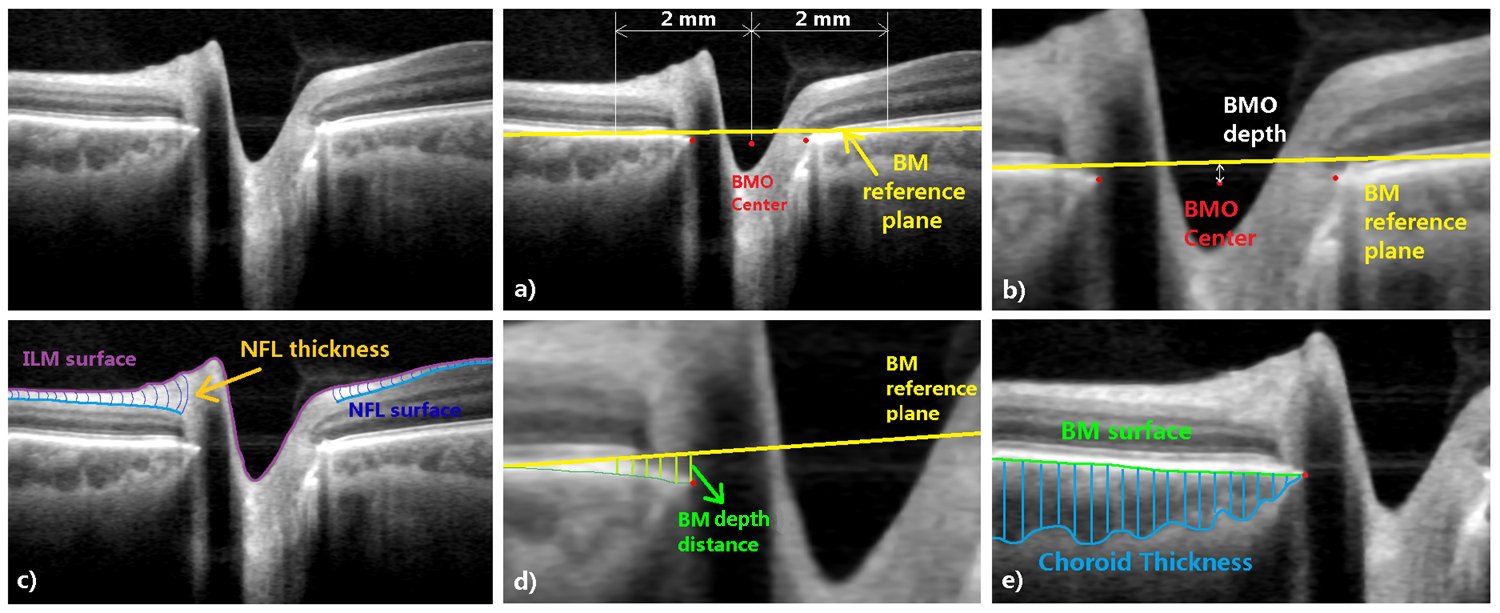
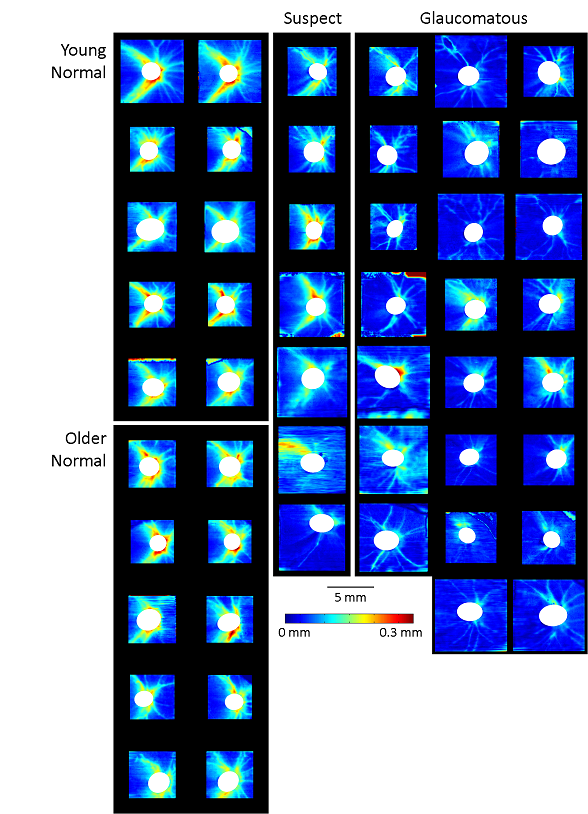 Publications
Publications