Overview
Our lab is studying neuronal development in C. elegans. We are focussing on the question of how neuronal circuits are formed. Projects in the lab currently use various complementary approaches to identify and study genes involved in pathfinding of neuronal processes (axon guidance).
Screens for axon guidance mutants
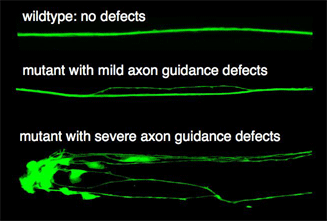
We use different approaches to identify genes controlling axon navigation. In genetic screens animals are mutagenized, which randomly destroys genes. Progeny with defective axon navigation can be identified in live animals when neurons are labelled with fluorescent markers like the green fluorescent protein (GFP). We collected a number of mutants in novel genes with defects ranging from subtle fasciculation defects within the ventral cord to severe defects affecting axon navigation in various parts of the nervous system (see Figure). The detailed characterization of those genes will lead to new insights into the genetic control of axon outgrowth. In a complementary approach we use double stranded RNA mediated gene silencing (RNAi) in large-scale screens to identify novel axon guidance genes. More recently we identified putative axon guidance receptors in a candidate gene approach examining mutants in genes encoding putative transmembrane proteins expressed in the developing nervous system.
The role of pioneer neurons in axon guidance
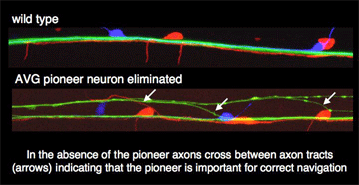
Neuronal processes do not grow out simultaneously from all neurons. There is a distinct set of neurons, pioneers, who are the first to send out their axons. These form the starting point for the major axon tracts and provide a first scaffold of pathways that are populated by all the later outgrowing axons (followers). It is thought that the pioneer axons provide crucial navigation information for later outgrowing axons. This idea can be easily tested in C. elegans, where individual cells can be eliminated by a method termed 'laser ablation'. In the absence of certain pioneer neurons, later outgrowing axons indeed commit navigation errors (see Figure), but they are not completely lost and many of them still find their way to their targets. In other words pioneer neurons do not seem to be absolutely essential, but provide one (or more) of many different navigation cues for later outgrowing axons. Recently we identified a member of the cadherin family as essential for pioneer-mediated navigation of follower axons (see next section) and have identified mutations essential for the correct navigation of the pioneer axons in the ventral nerve cord.
Cadherins in axon guidance
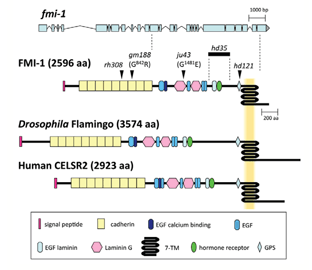
We found that two evolutionary conserved members of the cadherin family are essential for correct axonal navigation. cdh-4 mutant animals show fasciculation defects in the major longitudinal axon tracts, the ventral and dorsal nerve cords (Dev.Biol. 316(2):249). In addition these mutants have morphological defets, which are probably caused by adhesion defects in epidermal cells. The flamingo-type cadherin FMI-1 (see Figure) is required for the correct navigation of follower axons, which normally extend along the axons of earlier outgrowing pioneers (Development 137(21):3663). Both cadherins are likely to act as adhesion molecules. Currently we are investigating the molecular basis for their action and are in the process of identifying genes acting together with these cadherins.