Background
The following notation is used to describe the classification methods:
- x
- a p-dimensional vector containing the
quantitative variables of an observation
- Sp
- the pooled covariance matrix
- t
- a subscript to distinguish the groups
- nt
- the number of training set observations in group t
- mt
- the p-dimensional vector containing variable means in group t
- St
- the covariance matrix within group t

- the determinant of St
- qt
- the prior probability of membership in group t

- the posterior probability of an observation
x belonging to group t
- ft
- the probability density function for group t
- ft(x)
- the group-specific density estimate
at x from group t
- f(x)
 , the estimated
unconditional density at x
, the estimated
unconditional density at x
- et
- the classification error rate for group t
Bayes' Theorem
Assuming that the prior probabilities of group membership are
known and that the group-specific densities at x can be
estimated, PROC DISCRIM computes  , the probability of
x belonging to group t, by applying Bayes' theorem:
, the probability of
x belonging to group t, by applying Bayes' theorem:

PROC DISCRIM partitions a p-dimensional vector space into
regions Rt, where the region Rt is the subspace
containing all p-dimensional vectors y such
that  is the largest among all groups.
An observation is classified as coming
from group t if it lies in region Rt.
is the largest among all groups.
An observation is classified as coming
from group t if it lies in region Rt.
Parametric Methods
Assuming that each group has a multivariate normal distribution,
PROC DISCRIM develops a discriminant function or classification
criterion using a measure of generalized squared distance.
The classification criterion is based on either
the individual within-group covariance matrices
or the pooled covariance matrix; it also takes
into account the prior probabilities of the classes.
Each observation is placed in the class from which
it has the smallest generalized squared distance.
PROC DISCRIM also computes the posterior probability
of an observation belonging to each class.
The squared Mahalanobis distance from x to group t is
-
dt2(x) = (x - mt)' Vt-1(x - mt)
where Vt = St if the within-group
covariance matrices are used, or Vt = Sp
if the pooled covariance matrix is used.
The group-specific density estimate at
x from group t is then given by

Using Bayes' theorem, the posterior probability
of x belonging to group t is

where the summation is over all groups.
The generalized squared distance from
x to group t is defined as
-
Dt2(x) = dt2(x) + g1(t) + g2(t)
where

and

The posterior probability of x
belonging to group t is then equal to

The discriminant scores are -0.5 Du2(x).
An observation is classified into group u if
setting t=u produces the largest value of
 or the smallest value of Dt2(x).
If this largest posterior probability is less than the
threshold specified, x is classified into group OTHER.
or the smallest value of Dt2(x).
If this largest posterior probability is less than the
threshold specified, x is classified into group OTHER.
Nonparametric discriminant methods are based on nonparametric
estimates of group-specific probability densities.
Either a kernel method or the k-nearest-neighbor method
can be used to generate a nonparametric density estimate
in each group and to produce a classification criterion.
The kernel method uses uniform, normal, Epanechnikov,
biweight, or triweight kernels in the density estimation.
Either Mahalanobis distance or Euclidean
distance can be used to determine proximity.
When the k-nearest-neighbor method is used, the Mahalanobis
distances are based on the pooled covariance matrix.
When a kernel method is used, the Mahalanobis distances
are based on either the individual within-group
covariance matrices or the pooled covariance matrix.
Either the full covariance matrix or the diagonal matrix of
variances can be used to calculate the Mahalanobis distances.
The squared distance between two observation vectors,
x and y, in group t is given by
-
dt2(x,y) = (x-y)' Vt-1 (x-y)
where Vt has one of the following forms:

The classification of an observation vector x is based
on the estimated group-specific densities from the training set.
From these estimated densities, the posterior probabilities
of group membership at x are evaluated.
An observation x is classified into group u if
setting t=u produces the largest value of  .If there is a tie for the largest probability or
if this largest probability is less than the threshold
specified, x is classified into group OTHER.
.If there is a tie for the largest probability or
if this largest probability is less than the threshold
specified, x is classified into group OTHER.
The kernel method uses a fixed radius, r, and a
specified kernel, Kt, to estimate the group t
density at each observation vector x.
Let z be a p-dimensional vector.
Then the volume of a p-dimensional unit sphere
bounded by z'z = 1 is

where  represents the gamma function (refer to
SAS Language Reference: Dictionary).
represents the gamma function (refer to
SAS Language Reference: Dictionary).
Thus, in group t, the volume of a
p-dimensional ellipsoid bounded by
 is
is

The kernel method uses one of the following
densities as the kernel density in group t.
Uniform Kernel

Normal Kernel (with mean zero, variance r2 Vt)
-
Kt(z) = [1/(c0(t))] exp(-[1/(2r2)] z' Vt-1 z )
where  .
.
Epanechnikov Kernel

where
c1(t) = [1/(vr(t))] ( 1 + [p/2] ).
Biweight Kernel

where
c2(t) = (1 + [p/4] ) c1(t).
Triweight Kernel

where
c3(t) = ( 1 + [p/6] ) c2(t).
The group t density at x is estimated by

where the summation is over all observations y
in group t, and Kt is the specified kernel function.
The posterior probability of membership
in group t is then given by

where  is the estimated unconditional density.
If f(x) is zero, the observation
x is classified into group OTHER.
is the estimated unconditional density.
If f(x) is zero, the observation
x is classified into group OTHER.
The uniform-kernel method treats Kt(z) as a multivariate
uniform function with density uniformly distributed over
 .Let kt be the number of training set observations y
from group t within the closed ellipsoid centered at
x specified by
.Let kt be the number of training set observations y
from group t within the closed ellipsoid centered at
x specified by 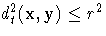 .Then the group t density at x is estimated by
.Then the group t density at x is estimated by
-
ft(x) = [(kt)/(nt vr(t))]
When the identity matrix or the pooled within-group
covariance matrix is used in calculating the squared distance,
vr(t) is a constant, independent of group membership.
The posterior probability of x
belonging to group t is then given by

If the closed ellipsoid centered at x does not
include any training set observations, f(x)
is zero and x is classified into group OTHER.
When the prior probabilities are equal,  is proportional to kt/nt and x is classified
into the group that has the highest proportion
of observations in the closed ellipsoid.
When the prior probabilities are proportional to
the group sizes,
is proportional to kt/nt and x is classified
into the group that has the highest proportion
of observations in the closed ellipsoid.
When the prior probabilities are proportional to
the group sizes,  ,
x is classified into the group that has the
largest number of observations in the closed ellipsoid.
,
x is classified into the group that has the
largest number of observations in the closed ellipsoid.
The nearest-neighbor method fixes the number, k,
of training set points for each observation x.
The method finds the radius rk(x) that is
the distance from x to the kth nearest
training set point in the metric Vt-1.
Consider a closed ellipsoid centered at x bounded by
 ; the nearest-neighbor
method is equivalent to the uniform-kernel method
with a location-dependent radius rk(x).
Note that, with ties, more than k training set points may
be in the ellipsoid.
; the nearest-neighbor
method is equivalent to the uniform-kernel method
with a location-dependent radius rk(x).
Note that, with ties, more than k training set points may
be in the ellipsoid.
Using the k-nearest-neighbor rule,
the kn (or more with ties) smallest distances are saved.
Of these k distances, let kt represent the number
of distances that are associated with group t.
Then, as in the uniform-kernel method, the
estimated group t density at x is
-
ft(x) = [(kt)/(nt vk(x))]
where vk(x) is the volume of the ellipsoid
bounded by  .Since the pooled within-group covariance matrix
is used to calculate the distances used in the
nearest-neighbor method, the volume vk(x)
is a constant independent of group membership.
When k=1 is used in the nearest-neighbor rule,
x is classified into the group associated
with the y point that yields the smallest
squared distance dt2(x,y).
Prior probabilities affect nearest-neighbor results in the same way
that they affect uniform-kernel results.
.Since the pooled within-group covariance matrix
is used to calculate the distances used in the
nearest-neighbor method, the volume vk(x)
is a constant independent of group membership.
When k=1 is used in the nearest-neighbor rule,
x is classified into the group associated
with the y point that yields the smallest
squared distance dt2(x,y).
Prior probabilities affect nearest-neighbor results in the same way
that they affect uniform-kernel results.
With a specified squared distance formula (METRIC=,
POOL=), the values of r and k determine the
degree of irregularity in the estimate of the density
function, and they are called smoothing parameters.
Small values of r or k produce jagged density estimates, and
large values of r or k produce smoother density estimates.
Various methods for choosing the smoothing parameters have been
suggested, and there is as yet no simple solution to this problem.
For a fixed kernel shape, one way to choose the smoothing
parameter r is to plot estimated densities with different
values of r and to choose the estimate that is most in
accordance with the prior information about the density.
For many applications, this approach is satisfactory.
Another way of selecting the smoothing parameter r
is to choose a value that optimizes a given criterion.
Different groups may have different sets of optimal values.
Assume that the unknown density has bounded and
continuous second derivatives and that the kernel
is a symmetric probability density function.
One criterion is to minimize an approximate mean integrated
square error of the estimated density (Rosenblatt 1956).
The resulting optimal value of r depends
on the density function and the kernel.
A reasonable choice for the smoothing parameter r is to
optimize the criterion with the assumption that group t
has a normal distribution with covariance matrix Vt.
Then, in group t, the resulting
optimal value for r is given by
-
( [(A(Kt))/(nt)] )[1/(p+4)]
where the optimal constant A(Kt) depends
on the kernel Kt (Epanechnikov 1969).
For some useful kernels, the constants A(Kt) are given by

These selections of A(Kt) are derived under the
assumption that the data in each group are from a multivariate
normal distribution with covariance matrix Vt.
However, when the Euclidean distances are used in
calculating the squared distance
(Vt = I), the smoothing constant should be
multiplied by s,
where s is an estimate of standard deviations for all variables.
A reasonable choice for s is

where sjj are group t marginal variances.
The DISCRIM procedure uses only a single smoothing parameter for all groups.
However, with the selection of the matrix to be used in the
distance formula (using the METRIC= or POOL= option),
individual groups and variables can have different scalings.
When Vt, the matrix used in calculating the squared
distances, is an identity matrix, the kernel estimate on each
data point is scaled equally for all variables in all groups.
When Vt is the diagonal matrix of a covariance matrix,
each variable in group t is scaled separately by its variance
in the kernel estimation, where the variance can be the pooled
variance (Vt = Sp) or an individual within-group
variance (Vt = St).
When Vt is a full covariance matrix, the
variables in group t are scaled simultaneously
by Vt in the kernel estimation.
In nearest-neighbor methods, the choice of k
is usually relatively uncritical (Hand 1982).
A practical approach is to try several different values
of the smoothing parameters within the context of the
particular application and to choose the one that gives
the best cross validated estimate of the error rate.
Classification Error-Rate Estimates
A classification criterion can be evaluated by its
performance in the classification of future observations.
PROC DISCRIM uses two types of error-rate estimates to
evaluate the derived classification criterion based
on parameters estimated by the training sample:
- error-count estimates
- posterior probability error-rate estimates.
The error-count estimate is calculated by applying
the classification criterion derived from the
training sample to a test set and then counting
the number of misclassified observations.
The group-specific error-count estimate is the
proportion of misclassified observations in the group.
When the test set is independent of the
training sample, the estimate is unbiased.
However, it can have a large variance,
especially if the test set is small.
When the input data set is an ordinary SAS data set and no
independent test sets are available, the same data set can be
used both to define and to evaluate the classification criterion.
The resulting error-count estimate has an optimistic
bias and is called an apparent error rate.
To reduce the bias, you can split the data into two
sets, one set for deriving the discriminant function
and the other set for estimating the error rate.
Such a split-sample method has the unfortunate
effect of reducing the effective sample size.
Another way to reduce bias is cross validation (Lachenbruch
and Mickey 1968).
Cross validation treats n-1 out of n
training observations as a training set.
It determines the discriminant functions based
on these n-1 observations and then applies
them to classify the one observation left out.
This is done for each of the n training observations.
The misclassification rate for each group is the proportion
of sample observations in that group that are misclassified.
This method achieves a nearly unbiased
estimate but with a relatively large variance.
To reduce the variance in an error-count estimate,
smoothed error-rate estimates are suggested (Glick 1978).
Instead of summing terms that are either zero or one as in the
error-count estimator, the smoothed estimator uses a continuum
of values between zero and one in the terms that are summed.
The resulting estimator has a smaller
variance than the error-count estimate.
The posterior probability error-rate estimates provided by
the POSTERR option in the PROC DISCRIM statement (see the
following section,
"Posterior Probability Error-Rate Estimates")
are smoothed error-rate estimates.
The posterior probability estimates for each group
are based on the posterior probabilities of the
observations classified into that same group.
The posterior probability estimates provide good estimates of
the error rate when the posterior probabilities are accurate.
When a parametric classification criterion (linear
or quadratic discriminant function) is derived from
a nonnormal population, the resulting posterior
probability error-rate estimators may not be appropriate.
The overall error rate is estimated through a weighted
average of the individual group-specific error-rate estimates,
where the prior probabilities are used as the weights.
To reduce both the bias and the variance of the
estimator, Hora and Wilcox (1982) compute the posterior
probability estimates based on cross validation.
The resulting estimates are intended to have both
low variance from using the posterior probability
estimate and low bias from cross validation.
They use Monte Carlo studies on two-group multivariate
normal distributions to compare the cross validation
posterior probability estimates with three other
estimators: the apparent error rate, cross validation
estimator, and posterior probability estimator.
They conclude that the cross validation posterior probability
estimator has a lower mean squared error in their simulations.
Consider the plot shown in Figure 25.6 with two variables,
X1 and X2, and two classes, A and B. The within-class
covariance matrix is diagonal, with a positive value for X1 but
zero for X2. Using a Moore-Penrose pseudo-inverse would
effectively ignore X2 completely in doing the classification,
and the two classes would have a zero generalized distance and could
not be discriminated at all. The quasi-inverse used by PROC DISCRIM
replaces the zero variance for X2 by a small positive number to
remove the singularity. This allows X2 to be used in the
discrimination and results correctly in a large generalized distance
between the two classes and a zero error rate. It also allows new
observations, such as the one indicated by N, to be classified in a
reasonable way. PROC CANDISC also uses a quasi-inverse when the
total-sample covariance matrix is considered to be singular and
Mahalanobis distances are requested. This problem with singular
within-class covariance matrices is discussed in Ripley (1996,
p. 38). The use of the quasi-inverse is an innovation introduced by
SAS Institute Inc.
Figure 25.6: Plot of Data with Singular Within-Class Covariance Matrix
Let S be a singular covariance matrix. The matrix S can be
either a within-group covariance matrix, a pooled covariance matrix,
or a total-sample covariance matrix. Let v be the number of
variables in the VAR statement and the nullity n be the number of
variables among them with (partial) R2 exceeding 1-p. If the
determinant of S (Testing of Homogeneity of Within Covariance
Matrices) or the inverse of S (Squared Distances and
Generalized Squared Distances) is required, a quasi-determinant or
quasi-inverse is used instead. PROC DISCRIM scales each variable
to unit total-sample variance before calculating this quasi-inverse.
The calculation is based on the spectral decomposition  , where
, where  is a diagonal
matrix of eigenvalues
is a diagonal
matrix of eigenvalues  , j = 1, ... , v, where
, j = 1, ... , v, where
 when i<j, and
when i<j, and  is a matrix
with the corresponding orthonormal eigenvectors of S as
columns. When the nullity n is less than v, set
is a matrix
with the corresponding orthonormal eigenvectors of S as
columns. When the nullity n is less than v, set  for j = 1, ... , v-n, and
for j = 1, ... , v-n, and  for j = v-n+1, ... , v, where
for j = v-n+1, ... , v, where

When the nullity n is equal to v, set  , for j = 1, ... , v. A quasi-determinant is then defined as the product of
, for j = 1, ... , v. A quasi-determinant is then defined as the product of
 , j = 1, ... , v. Similarly, a quasi-inverse is
then defined as
, j = 1, ... , v. Similarly, a quasi-inverse is
then defined as  , where
, where  is a diagonal matrix of
values
is a diagonal matrix of
values  j = 1, ... , v.
j = 1, ... , v.
Copyright © 1999 by SAS Institute Inc., Cary, NC, USA. All rights reserved.



















