STAT 350: Lecture 32
Example: transformation vs glm
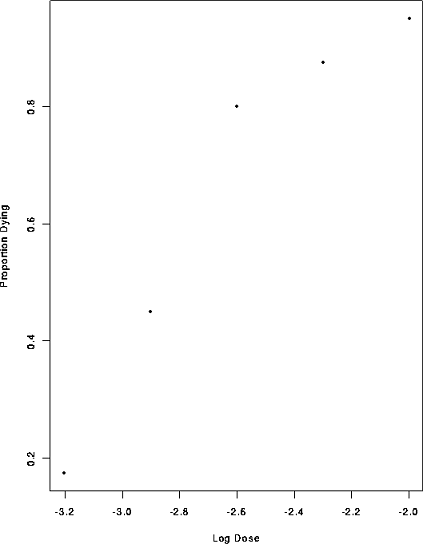
At each of 5 doses of some drug 40 animals were tested and the number dying were recorded. The log doses are -3.204, -2.903, -2.602, -2.301, and -2.000 and the numbers surviving are 7, 18, 32, 35, 38. A plot of the data is

![]()

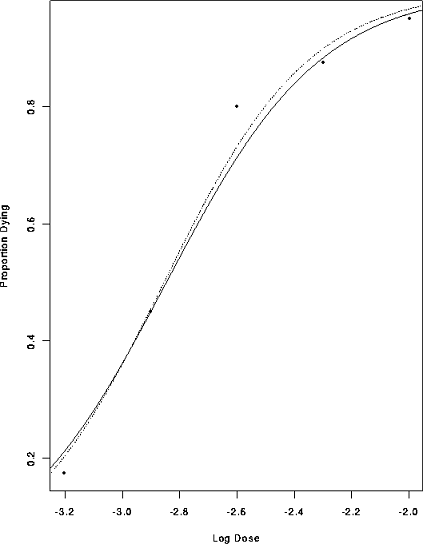
Here is the data analyzed by least squares after the logistic transformation
THE DATA FILE
Dose Dead Tested -3.204 7 40 -2.903 18 40 -2.602 32 40 -2.301 35 40 -2.000 38 40
I used SPlus to analyze the data; here is a file full of commands I used.
dead <- read.table("data", header = T)
attach(dead)
postscript("logist.ps",onefile=F,horizontal=F)
plot(Dose, Dead/Tested,xlab="Log Dose",ylab="Proportion Dying")
dev.off()
postscript("arc_logist.ps",onefile=F,horizontal=F)
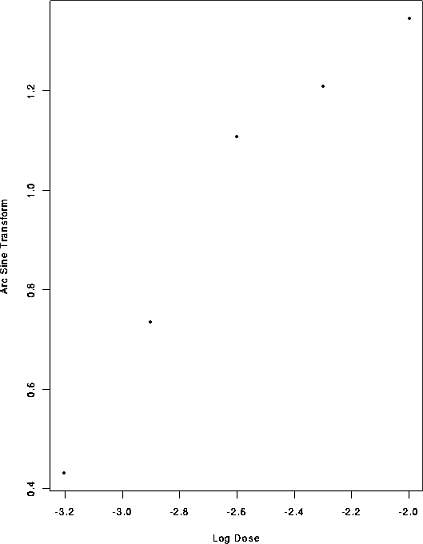
plot(Dose, Dead/Tested,xlab="Log Dose",ylab="Arc Sine Transform")
dev.off()
postscript("logit_logist.ps",onefile=F,horizontal=F)
plot(Dose, log(Dead/(Tested-Dead)),xlab="Log Dose",ylab="Logit Transform")
dev.off()
linfit <- lm( log(Dead/(Tested-Dead)) ~ Dose, data=dead)
summary(linfit)
glmfit <- glm( cbind(Dead,Tested-Dead) ~ Dose, data=dead, family=binomial)
summary(glmfit)
postscript("logist_plus_curve.ps",onefile=F,horizontal=F)
plot(Dose, Dead/Tested,xlab="Log Dose",ylab="Proportion Dying")
d <- seq(-3.3,-1.9,length=200)
etalin <- coef(linfit)[1] + d*coef(linfit)[2]
p <- exp(etalin)/(1+exp(etalin))
lines(d,p)
etaglm <- coef(glmfit)[1] + d*coef(glmfit)[2]
p <- exp(etaglm)/(1+exp(etaglm))
lines(d,p,lty=2)
dev.off()
The output consists of 4 postscript files which have graphs in them and the following
printout:
S-PLUS : Copyright (c) 1988, 1996 MathSoft, Inc.
S : Copyright AT&T.
Version 3.4 Release 1 for Sun SPARC, SunOS 5.3 : 1996
Working data will be in .Data
> dead <- read.table("data", header = T)
#
# Read in the data. Columns are named by the words
# read off line 1 because of the header=T bit.
#
> attach(dead)
#
# Makes the variable which are columns of dead
# accessible to the plotting rooutines
#
> postscript("logist.ps",onefile=F,horizontal=F)
#
# Declares that the next graph should be put
# in a postscript file called logist.ps. The
# file should be encapsulated postscript and in
# portrait orientation.
#
> plot(Dose, Dead/Tested,xlab="Log Dose",ylab="Proportion Dying")
#
# Plot Proportion dying on the y axis against Dose and
# label the axes
#
> dev.off()
#
# Finish up the postscript file
#
Starting to make postscript file.
Finished postscript file, executing command "lpr -h logist.ps &".
null device
1
> postscript("arc_logist.ps",onefile=F,horizontal=F)
> plot(Dose, asin(sqrt(Dead/Tested)),xlab="Log Dose",ylab="Arc Sine Transform")
> dev.off()
Starting to make postscript file.
Finished postscript file, executing command "lpr -h arc_logist.ps &".
null device
1
> postscript("logit_logist.ps",onefile=F,horizontal=F)
> plot(Dose, log(Dead/(Tested-Dead)),xlab="Log Dose",ylab="Logit Transform")
> dev.off()
Starting to make postscript file.
Finished postscript file, executing command "lpr -h logit_logist.ps &".
null device
1
> > linfit <- lm( log(Dead/(Tested-Dead)) ~ Dose, data=dead)
#
# Regress log(Y/(n-Y)) on Dose
#
> summary(linfit)
#
# Print out a summary of the regression results.
#
Call: lm(formula = log(Dead/(Tested - Dead)) ~ Dose, data = dead)
Residuals:
1 2 3 4 5
-0.2283 0.00792 0.4812 -0.07283 -0.188
Coefficients:
Value Std. Error t value Pr(>|t|)
(Intercept) 10.5322 0.9109 11.5626 0.0014
Dose 3.6999 0.3455 10.7095 0.0017
Residual standard error: 0.3288 on 3 degrees of freedom
Multiple R-Squared: 0.9745
F-statistic: 114.7 on 1 and 3 degrees of freedom, the p-value is 0.001741
Correlation of Coefficients:
(Intercept)
Dose 0.9869
> glmfit <- glm( cbind(Dead,Tested-Dead) ~ Dose, data=dead, family=binomial)
#
# This fits the model that log(E(Y)/(n-E(Y)) is a linear function
# of Dose
#
> summary(glmfit)
Call: glm(formula = cbind(Dead, Tested - Dead) ~ Dose, family = binomial, data = dead
)
Deviance Residuals:
1 2 3 4 5
-0.4319303 -0.03563685 1.026423 -0.476305 -0.5453582
Coefficients:
Value Std. Error t value
(Intercept) 11.238232 1.5651480 7.180300
Dose 3.936472 0.5604985 7.023162
(Dispersion Parameter for Binomial family taken to be 1 )
Null Deviance: 80.77441 on 4 degrees of freedom
Residual Deviance: 1.76566 on 3 degrees of freedom
Number of Fisher Scoring Iterations: 3
Correlation of Coefficients:
(Intercept)
Dose 0.9929832
> > > postscript("logist_plus_curve.ps",onefile=F,horizontal=F)
> plot(Dose, Dead/Tested,xlab="Log Dose",ylab="Proportion Dying")
> d <- seq(-3.3,-1.9,length=200)
> etalin <- coef(linfit)[1] + d*coef(linfit)[2]
> p <- exp(etalin)/(1+exp(etalin))
#
# For each dose in d, which is a list of 200 numbers running
# from -3.3 to -1.9, compute the fitted probability according to
# the logit model: if log(x/(1-x))=p then p=exp(x)/(1+exp(x))
#
> lines(d,p)
#
# Plot the fitted curve for the least squares method on the
# graph of the data
#
> etaglm <- coef(glmfit)[1] + d*coef(glmfit)[2]
> p <- exp(etaglm)/(1+exp(etaglm))
#
# Do the same for the generalized model fit
#
> lines(d,p,lty=2)
#
# Plot the fitted curve for the glm method on the
# graph of the data. Use a dashed (lty=2) line.
#
> dev.off()
Starting to make postscript file.
Finished postscript file, executing command "lpr -h logist_plus_curve.ps &".
null device
1
> q() # end-of-file
Poisson Regression: Count Data
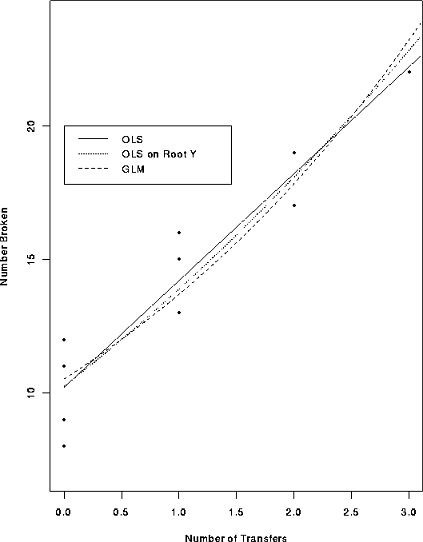
In the table below the first row is the number of times a carton of glass objects was transferred from one aircraft to another during shipping. The second row is the number of broken objects.
| i: | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 |
|
| 1 | 0 | 2 | 0 | 3 | 1 | 0 | 1 | 2 | 0 |
|
| 16 | 9 | 17 | 12 | 22 | 13 | 8 | 15 | 19 | 11 |
A reasonable model is that ![]() has a Poisson distribution with
mean
has a Poisson distribution with
mean ![]() which depends in some way on
which depends in some way on ![]() . We fit 3 models:
. We fit 3 models:
![]()
for which the relevant plot is
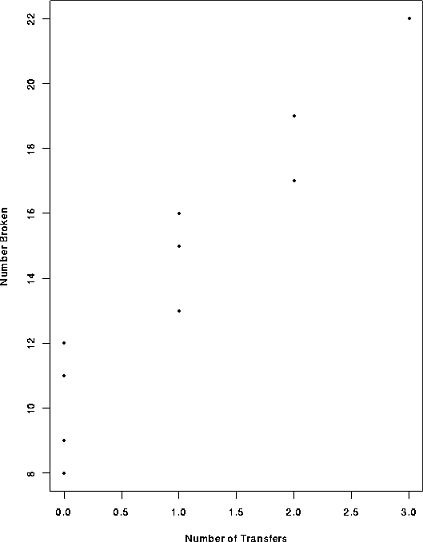
![]()
for which the relevant plot is

![]()
or equivalently
![]()
The following SPlus code fits these models
dat <- read.table("data", header = T)
attach(dat)
postscript("xyplot.ps",onefile=F,horizontal=F)
plot(Transfers,Broken,xlab="Number of Transfers",ylab="Number Broken")
dev.off()
postscript("xrootyplot.ps",onefile=F,horizontal=F)
plot(Transfers, sqrt(Broken),xlab="Number of Transfers",ylab="Square Root Transform")
dev.off()
#
# Regress Number Broken on Number of Transfers
#
linfit <- lm( Broken ~ Transfers, data=dat)
summary(linfit)
diag(linfit)
#
# Regress Square Root of Number Broken on Number of Transfers
#
rootlinfit <- lm( sqrt(Broken) ~ Transfers, data=dat)
summary(rootlinfit)
diag(rootlinfit)
#
# The following fits log(E(Y)) is a linear function
# of Dose and variance is equal to the mean
#
glmfit <- glm( Broken ~ Transfers, data=dat, family=Poisson)
summary(glmfit)
postscript("points_plus_curve.ps",onefile=F,horizontal=F)
plot(Transfers,Broken,xlab="Number of Transfers",ylab="Number Broken")
d <- seq(0,4,length=200)
etalin <- coef(linfit)[1] + d*coef(linfit)[2]
lines(d,etalin)
etarootlin <- coef(rootlinfit)[1] + d*coef(rootlinfit)[2]
lines(d,etarootlin^2,lty=2)
etaglm <- coef(glmfit)[1] + d*coef(glmfit)[2]
p <- exp(etaglm)
lines(d,p,lty=3)
legend(0,20,lty=1:3,legend=c("OLS","OLS on Root Y","GLM"))
dev.off()
EDITED OUTPUT (Complete output.)
> summary(linfit)
Call: lm(formula = Broken ~ Transfers, data = dat)
Residuals:
Min 1Q Median 3Q Max
-2.2 -1.2 0.3 0.8 1.8
Coefficients:
Value Std. Error t value Pr(>|t|)
(Intercept) 10.2000 0.6633 15.3771 0.0000
Transfers 4.0000 0.4690 8.5280 0.0000
Residual standard error: 1.483 on 8 degrees of freedom
Multiple R-Squared: 0.9009
F-statistic: 72.73 on 1 and 8 degrees of freedom, the p-value is 2.749e-05
Correlation of Coefficients:
(Intercept)
Transfers -0.7071
> summary(rootlinfit)
Call: lm(formula = sqrt(Broken) ~ Transfers, data = dat)
Residuals:
Min 1Q Median 3Q Max
-0.3722 -0.1263 0.01059 0.1392 0.274
Coefficients:
Value Std. Error t value Pr(>|t|)
(Intercept) 3.2006 0.1010 31.6789 0.0000
Transfers 0.5254 0.0714 7.3538 0.0001
Residual standard error: 0.2259 on 8 degrees of freedom
Multiple R-Squared: 0.8711
F-statistic: 54.08 on 1 and 8 degrees of freedom, the p-value is 7.965e-05
Correlation of Coefficients:
(Intercept)
Transfers -0.7071
> summary(glmfit)
Call: glm(formula = Broken ~ Transfers, family = poisson, data = dat)
Deviance Residuals:
Min 1Q Median 3Q Max
-0.8105292 -0.2389281 -0.02029464 0.3299091 0.6074179
Coefficients:
Value Std. Error t value
(Intercept) 2.3529495 0.1317376 17.860883
Transfers 0.2638422 0.0792345 3.329891
(Dispersion Parameter for Poisson family taken to be 1 )
Null Deviance: 12.56868 on 9 degrees of freedom
Residual Deviance: 1.813176 on 8 degrees of freedom
Number of Fisher Scoring Iterations: 3
Correlation of Coefficients:
(Intercept)
Transfers -0.770864
The fits together