Research Topics
- Modern clinical trials and Bayesian inference
- Space-filling designs on constrained regions
- Informative priors and Bayesian computation
- Bayesian inference for social networks using aggregated relational data
- A Bayesian discovery procedure
- Sequentially constrained Monte Carlo
- Monotone emulation of computer experiments
Modern clinical trials and Bayesian inference
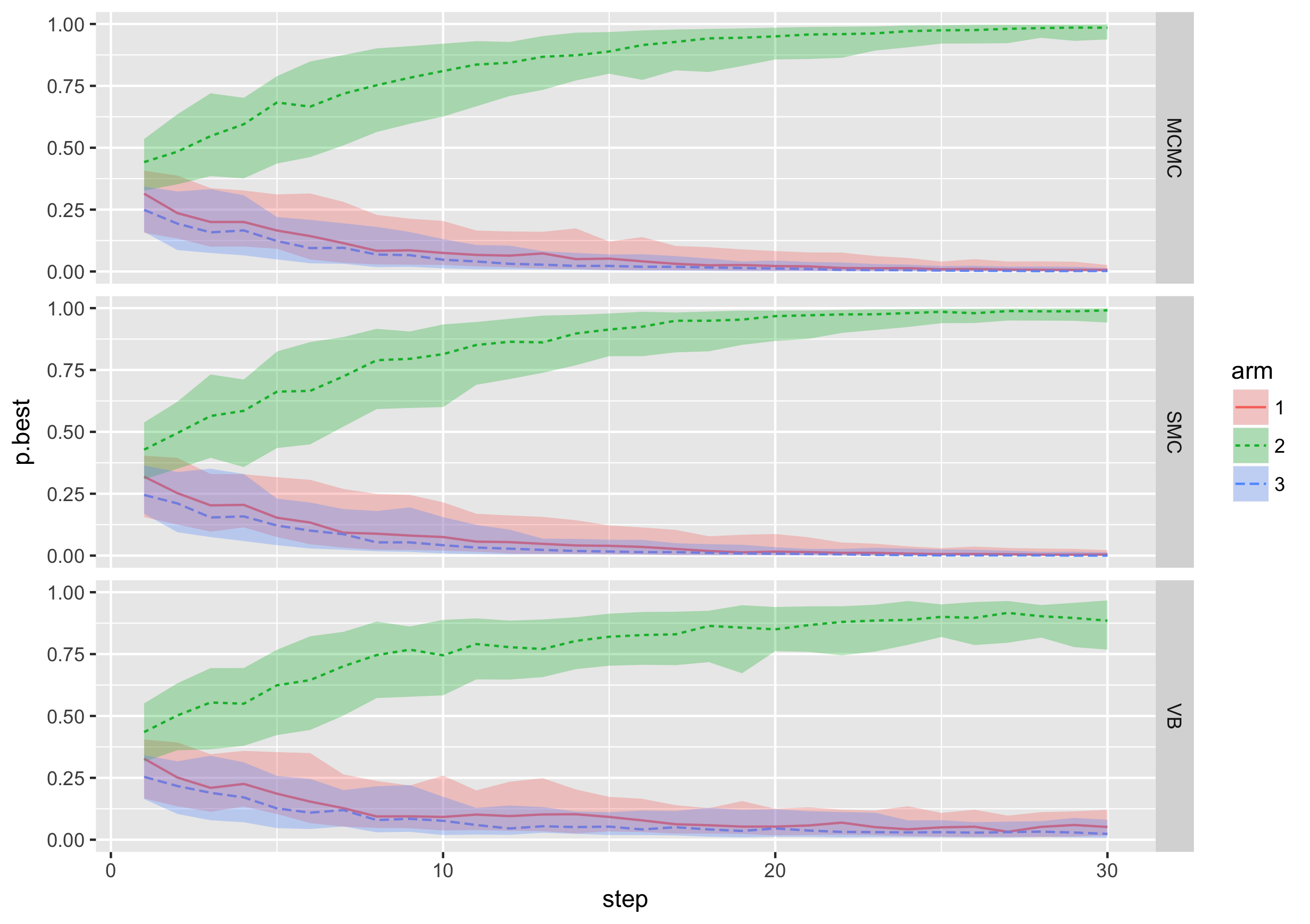
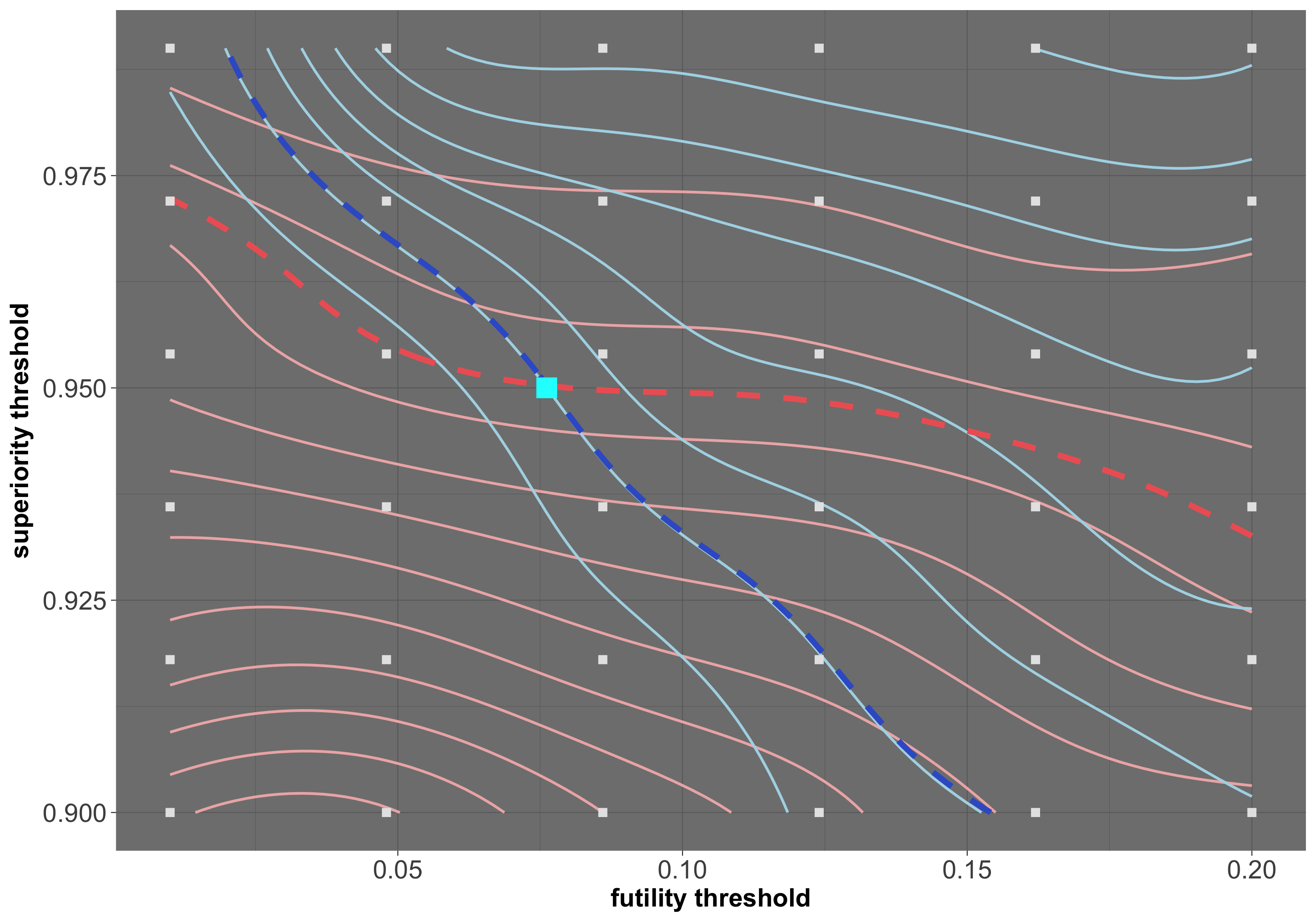
At MTEK Sciences we aim to contribute to global health and specifically child development by investigating, implementing and developing statistical methodology for design of clinical trials. Focusing on Bayesian adaptive designs, I leverage my experience in Bayesian modelling and computation to recommend efficient trial designs capable of answering important questions in complex settings.
Examples of my contributions to the field, so far, includes development of a Bayesian model for incorporating fractional additivity assumption for treatment combination trials (paper), tailoring an SMC algorithm for simulation of Bayesian adaptive trials (SMC.RAR R package) and Gaussian process emulation for power/error rate optimization in Bayesian adaptive designs (work in progress). In addition, I have developed a user-friendly shiny app for exploration of Bayesian adaptive designs through simulations -- give it a try here!
Space-filling designs on constrained regions

In many areas of science computer models are used instead of physical experiments to study complex systems. These models are often very expensive to run. This is where statistics plays an important role: a stochastic model is used to emulate the computer model based on a finite set of initial runs. However, the performance of the emulator depends on this initial runs. This has resulted in a sub-branch of design of experiments referred to as design of computer experiments that focuses on selecting an "optimal" set of inputs to run the compuetr model. Until recently design of computer experiments methodology was focused on rectangular input spaces, i.e., hypercubes. But in reality the inputs of computer models are often constrained. In many cases these constraints result in non-convex and highly irregular input spaces.
Many of the existing design construction methods can be generalized to be used on constrained regions if an initial uniformly covering sample is available over the input region. However, generating such a sample is not trivial for highly constrained and high-dimentional input spaces.
My contribution is a novel application of sequential Monte Carlo to generate a set of candidate design points on any arbitrarily constrained region. This is a specific application of sequentially Constrained Monte Carlo that my other paper is focused on. In a paper co-authored with Jason Loeppky, this sampling algorithm together with some Monte Carlo based design algorithms are introduced.
Informative priors and Bayesian computation
The use of prior distribution is often a controversial topic in Bayesian inference. Informative priors are often avoided at all costs. However, when prior information is available informative priors are an appropriate way of introducing this information into the model. Furthermore, informative priors, when used properly and creatively, can provide solutions to computational issues and improve modeling efficiency. In this paper I emphasize the usefulness of informative priors in a variety of different scenarios through three examples that I had encountered previously in my research.
Bayesian inference for social networks using aggregated relational data Aggregated relational data (ARD), i.e., data that are collected by asking respondents how many acquaintances they have in different sub-populations, can be used to make inference about the underlying social network. The existing Bayesian models for analysis of ARD (e.g. Zheng et al. (2006)) are computationally demanding and non-identifiable. The non-identifiability is dealth with by a renormalization step imbedded in the MCMC. The renormalization step takes advantage of external information about some sub-populations that is available from other sources such as the census or other social surveys. This computational complexity makes these model inaccessible to the users in social sciences.
During my post-doc at the Applied Statistics Centre at Columbia University I worked with
Tian Zheng and
Andrew Gelman to overcome these modelling issues. We propose to identify the model via an informative prior rather than an ad-hoc recentering step within the MCMC. This enables us to use the external information in a formal Bayesian framework and makes it possible for practitioners to implement the model using any Bayesian software such as Stan. The Stan implementation of the model and a brief discription of the model is available here.
A Bayesian discovery procedure
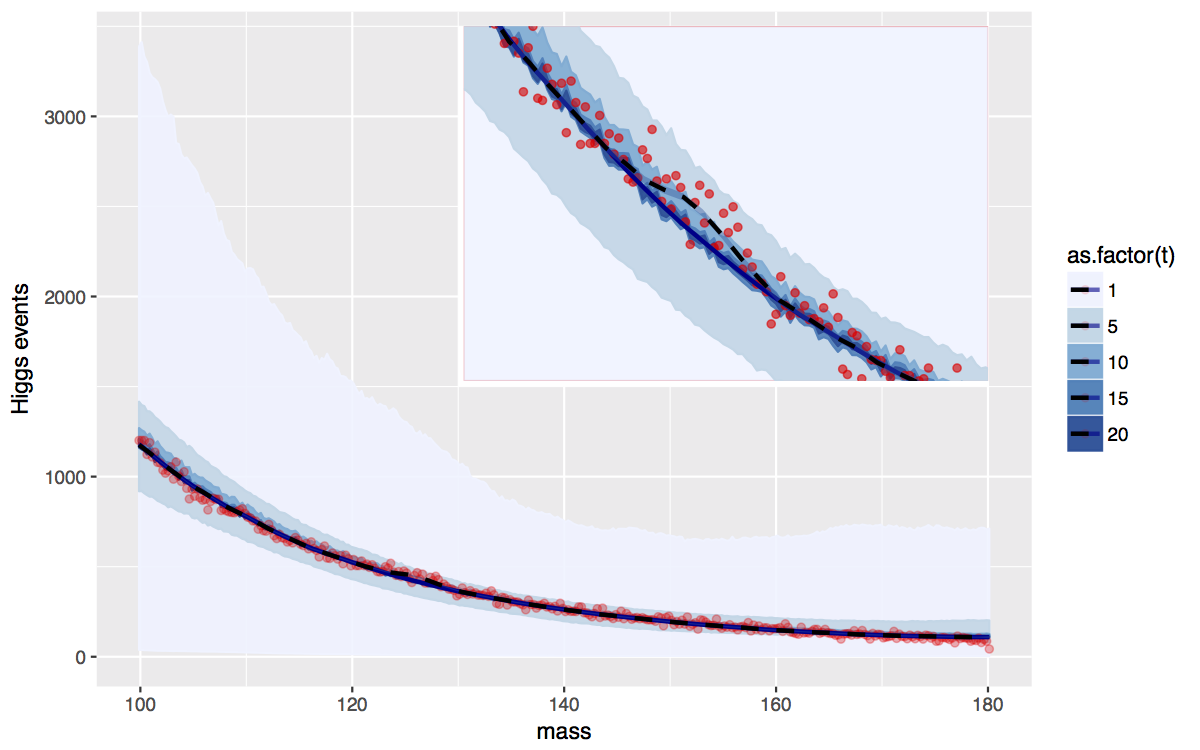
The Standard Model (SM) of particle physics is a theory that describes the dynamics of subatomic particles. The Higgs particle is an essential component of the SM; its existence explains why other elementary particles are massive. The existence of the Higgs boson needs to be confirmed by experiment. The Large Hadron Collider (LHC) at the European organization for nuclear research, known as CERN, is a high energy collider specifically designed and constructed to detect the Higgs particle. Two beams of protons circulating at very high speeds in the LHC collide inside two detectors (ATLAS and CMS). Collisions between a proton in one beam and a proton in the other beam result in generation of new particles, possibly including the Higgs boson; each such collision is an event. Some of the particles generated can be tracked and measured in the detectors. However, the Higgs particle, if generated, decays extremely quickly into other known SM particles and cannot be detected directly. Instead, the existence of the Higgs particle must be inferred by looking for those combinations of detectable particles that are predicted by the SM.
Once a Higgs particle has been created (by one of several "production mechanisms") in a
proton-proton collision, there are several different processes, called "decay modes", through
which the particle may decay. The decay process can be reconstructed based on the detected
collision byproducts. Events with reconstructed processes that match one of the
possible Higgs decay modes and pass other selection criteria (called cuts) are recorded as
"Higgs candidates" and the invariant mass of the unobserved particle is computed from the
reconstruction. A histogram of the estimator of the mass is then created for each decay mode.
However, there are other processes, not involving the Higgs boson, that can result in the
generation of Higgs event byproducts which also pass the cuts; these are called background
events. Thus the histogram created is either a mixture of background events and events in
which a Higgs particle was created or just a histogram of background events if the Higgs
particle does not exist.
In joint work with
Richard Lockhart we propose a Bayesian model as well as a Bayesian decision making procedure that incorporates the underlying assumptions provided by the theory and satisfies the strict frequency error rate requirements in particle phisics. The paper can be found
here.
Sequentially constrained Monte Carlo (SCMC)
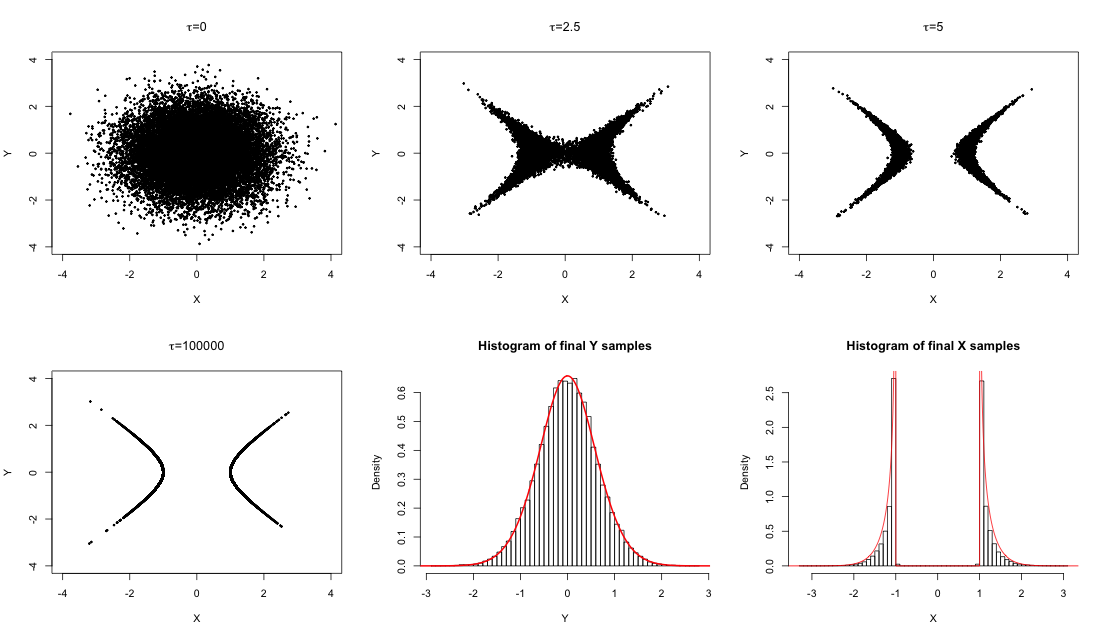
Constraints can be interpreted in a broad sense as any kind of explicit restriction over the parameters. While some constraints are defined directly on the parameter space, when they are instead defined by known behavior on the model, transformation of constraints into features on the parameter space may not be possible. Difficulties in sampling from the posterior distribution as a result of incorporation of constraints into the model is a common challenge leading to truncations in the parameter space and inefficient sampling algorithms. In joint work with
Dave Campbell we propose a variant of sequential Monte Carlo algorithm for posterior sampling in presence of constraints by defining a sequence of densities through the imposition of the constraint. Particles generated from an unconstrained or mildly constrained distribution are filtered and moved through sampling and resampling steps to obtain a sample from the fully constrained target distribution. General and model specific forms of constraints enforcing strategies are defined. The SCMC algorithm is demonstrated on constraints defined by monotonicity of a function, densities constrained to low dimensional manifolds, adherence to a theoretically derived model, and model feature matching. For more on SCMC see our paper.
Monotone emulation of computer experiments In statistical modeling of computer experiments, prior information is sometimes available about the underlying function. For example, the physical system simulated by the computer code may be known to be monotone with respect to some or all inputs. In joint work with
Derek Bingham,
Hugh Chipman, and
Dave Campbell we develop a Bayesian approach to Gaussian process modeling capable of incorporating monotonicity information for computer model emulation. Markov chain Monte Carlo methods are used to sample from the posterior distribution of the process given the simulator output and monotonicity information. The performance of the proposed approach in terms of predictive accuracy and uncertainty quantification is demonstrated in a number of simulated examples as well as a real queuing system application. See our paper here.