Estimation Methods
Consider the general nonlinear model:

where q is a real vector valued function, of
yt
is a real vector valued function, of
yt , xt
, xt ,
,
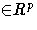 , g is the number of equations,
l is the number of exogenous
variables (lagged endogenous variables are considered exogenous here),
p is the number of parameters and t ranges from 1 to n.
zt
, g is the number of equations,
l is the number of exogenous
variables (lagged endogenous variables are considered exogenous here),
p is the number of parameters and t ranges from 1 to n.
zt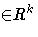 is a vector
of instruments.
is a vector
of instruments.  t is an unobservable disturbance
vector with the following properties:
t is an unobservable disturbance
vector with the following properties:

All of the methods implemented in PROC MODEL aim to minimize an
objective function. The following table summarizes the objective
functions defining the estimators and the corresponding
estimator of the covariance of the parameter estimates for each method.
Table 14.1: Summary of PROC MODEL Estimation Methods
|
Method
|
Instruments
|
Objective Function
|
Covariance of 
|
| OLS | no | r'r/n |  |
| ITOLS | no |  |  |
| SUR | no |  |  |
| ITSUR | no |  |  |
| N2SLS | yes |  |  |
| IT2SLS | yes |  |  |
| N3SLS | yes |  |  |
| IT3SLS | yes |  |  |
| GMM | yes | ![{[n{m}_{n}({\theta})]' \hat{V}\hspace*{1pt}^{-1}_{\rm{N2SLS}}[n{m}_{n}({\theta})]}](images/modeq43.gif) | ![{ [(Y{X})'\hat{V}^{-1}(Y{X})]^{-1}}](images/modeq44.gif) |
| ITGMM | yes | ![{[n{m}_{n}({\theta})]' \hat{V}\hspace*{1pt}^{-1}[n{m}_{n}({\theta})]}](images/modeq45.gif) | ![{ [(Y{X})'\hat{V}^{-1}(Y{X})]^{-1}}](images/modeq44.gif) |
| FIML | no | constant+[n/2]ln(det(S)) | ![{ [\hat{Z} ' ( S ^{-1} {\otimes} I) \hat{Z}]^{-1}}](images/modeq46.gif) |
| | |  | |
The column labeled "Instruments" identifies the estimation methods that
require instruments. The variables used in this table and the remainder of this
chapter are defined as follows:
n = is the number of nonmissing observations.
g = is the number of equations.
k = is the number of instrumental variables.
![{{r } = [\matrix{r_{1} \cr r_{2} \cr {\vdots} \cr r_{g}}] }](images/modeq48.gif) is the ng ×1 vector of residuals for the g equations stacked together.
is the ng ×1 vector of residuals for the g equations stacked together.
![{r_{i} = [\matrix{q_{i}(y_{1}\hspace*{1pt}, x_{1}\hspace*{1pt}, {{\theta}}) \cr ...
...) \cr {\vdots} \cr q_{i}(y_{n}\hspace*{1pt}, x_{n}\hspace*{1pt}, {{\theta}}) }]}](images/modeq49.gif) is the n ×1 column vector of residuals for the ith equation.
is the n ×1 column vector of residuals for the ith equation.
- S
-
is a g ×g matrix that estimates
 , the
covariances of the errors across equations (referred to as the S matrix).
, the
covariances of the errors across equations (referred to as the S matrix).
- X
- is an ng ×p matrix of partial derivatives of the residual with
respect to the parameters.
- W
- is an n ×n matrix, Z(Z'Z)-1Z'.
- Z
- is an n ×k matrix of instruments.
- Y
- is a gk ×ng matrix of instruments.
 .
.

 is an ng × p matrix.
is an ng × p matrix. 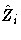 is a ng × 1
column vector obtained from stacking the columns of
is a ng × 1
column vector obtained from stacking the columns of

- U
- is an n × g matrix of residual errors.

- Q
- is the n × g matrix
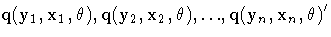 .
.
- Qi
- is an n × g matrix
 .
.
- I
- is an n ×n identity matrix.
- Jt
- is
 which is a g ×g Jacobian matrix.
which is a g ×g Jacobian matrix.
- mn
- is first moment of the crossproduct
 .
.

- zt
- is a k column vector of instruments for observation t. z't
is also the tth row of Z.

- is the gk ×gk matrix representing the variance of the moment functions.
- k
- is the number of instrumental variables used.
- constant
- is the constant
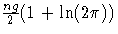 .
.

- is the notation for a Kronecker product.
All vectors are column vectors unless otherwise noted.
Other estimates of the covariance matrix for FIML are also available.
Dependent Regressors and Two-Stage Least Squares
Ordinary regression analysis is based on several assumptions.
A key assumption is that the independent variables are in fact
statistically independent of the unobserved error component of the model.
If this assumption is not true--if the regressor varies systematically
with the error--then ordinary regression produces inconsistent results.
The parameter estimates are biased.
Regressors might fail to be independent variables because they are dependent
variables in a larger simultaneous system.
For this reason, the problem of dependent regressors is
often called simultaneous equation bias.
For example, consider the following two-equation system.


In the first equation, y2 is a dependent, or endogenous, variable.
As shown by the second equation, y2 is a function of y1,
which by the first equation is a function of  1,
and therefore y2 depends on
1,
and therefore y2 depends on  1.
Likewise, y1 depends on
1.
Likewise, y1 depends on  2 and is a dependent regressor
in the second equation.
This is an example of a simultaneous equation system;
y1 and y2 are a function of all the variables in the system.
2 and is a dependent regressor
in the second equation.
This is an example of a simultaneous equation system;
y1 and y2 are a function of all the variables in the system.
Using the ordinary least squares (OLS) estimation method to estimate
these equations produces biased estimates.
One solution to this problem is to replace y1 and y2
on the right-hand side of the equations with predicted values,
thus changing the regression problem to the following:


This method requires estimating the predicted values
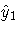 and
and 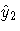 through a
preliminary, or "first stage,"
instrumental regression.
An instrumental regression is a regression of the dependent regressors
on a set of instrumental variables, which can be any independent variables useful for predicting the dependent regressors.
In this example, the equations are linear and the exogenous variables
for the whole system are known.
Thus, the best choice for instruments (of the variables in the model)
are the variables x1 and x2.
through a
preliminary, or "first stage,"
instrumental regression.
An instrumental regression is a regression of the dependent regressors
on a set of instrumental variables, which can be any independent variables useful for predicting the dependent regressors.
In this example, the equations are linear and the exogenous variables
for the whole system are known.
Thus, the best choice for instruments (of the variables in the model)
are the variables x1 and x2.
This method is known as two-stage least squares or 2SLS,
or more generally as the instrumental variables method.
The 2SLS method for linear models is discussed in Pindyck (1981, p. 191-192).
For nonlinear models this situation is more complex, but the idea is the same.
In nonlinear 2SLS, the derivatives of the model with respect to the parameters
are replaced with predicted values.
See the section "Choice of Instruments" for further
discussion of the use of
instrumental variables in nonlinear regression.
To perform nonlinear 2SLS estimation with PROC MODEL,
specify the instrumental variables with an INSTRUMENTS statement and
specify the 2SLS or N2SLS option on the FIT statement.
The following statements show how to estimate the first equation in
the preceding example with PROC MODEL.
proc model data=in;
y1 = a1 + b1 * y2 + c1 * x1;
fit y1 / 2sls;
instruments x1 x2;
run;
The 2SLS or instrumental variables estimator can be computed using a
first-stage regression on the instrumental variables as described previously.
However, PROC MODEL actually uses the equivalent but computationally more
appropriate technique of projecting the regression problem into the
linear space defined by the instruments.
Thus PROC MODEL does not produce any "first stage" results when you use 2SLS.
If you specify the FSRSQ option on the FIT statement,
PROC MODEL prints "first-stage R2" statistic for each parameter
estimate.
Formally, the  that minimizes
that minimizes

is the N2SLS estimator of the parameters. The estimate of  at the
final iteration is used in the covariance of the parameters
given in Table 14.1. Refer to Amemiya (1985, p. 250)
for details on the properties of nonlinear two-stage least squares.
at the
final iteration is used in the covariance of the parameters
given in Table 14.1. Refer to Amemiya (1985, p. 250)
for details on the properties of nonlinear two-stage least squares.
Seemingly Unrelated Regression
If the regression equations are not simultaneous, so there are
no dependent regressors, seemingly unrelated regression (SUR)
can be used to estimate systems of equations with correlated random errors.
The large-sample efficiency of an estimation can be improved
if these cross-equation correlations are taken into account.
SUR is also known as joint generalized least squares or
Zellner regression. Formally,
the  that minimizes
that minimizes

is the SUR estimator of the parameters.
The SUR method requires an estimate of the cross-equation covariance matrix,
 .PROC MODEL first performs an OLS estimation, computes
an estimate,
.PROC MODEL first performs an OLS estimation, computes
an estimate,  , from the OLS residuals,
and then performs the SUR estimation based on
, from the OLS residuals,
and then performs the SUR estimation based on  .The OLS results are not printed unless you specify the OLS option
in addition to the SUR option.
.The OLS results are not printed unless you specify the OLS option
in addition to the SUR option.
You can specify the  to use for SUR by storing
the matrix in a SAS data set and naming that data set
in the SDATA= option.
You can also feed the
to use for SUR by storing
the matrix in a SAS data set and naming that data set
in the SDATA= option.
You can also feed the  computed from the SUR residuals
back into the SUR estimation process by specifying the ITSUR option.
You can print the estimated covariance matrix
computed from the SUR residuals
back into the SUR estimation process by specifying the ITSUR option.
You can print the estimated covariance matrix  using the COVS option on the FIT statement.
using the COVS option on the FIT statement.
The SUR method requires estimation of the  matrix,
and this increases the sampling variability of the estimator
for small sample sizes.
The efficiency gain SUR has over OLS is a large sample property,
and you must have a reasonable amount of data to realize this gain.
For a more detailed discussion of SUR, refer to Pindyck (1981, p. 331-333).
matrix,
and this increases the sampling variability of the estimator
for small sample sizes.
The efficiency gain SUR has over OLS is a large sample property,
and you must have a reasonable amount of data to realize this gain.
For a more detailed discussion of SUR, refer to Pindyck (1981, p. 331-333).
Three-Stage Least-Squares Estimation
If the equation system is simultaneous, you can combine the 2SLS and SUR
methods to take into account both dependent regressors and
cross-equation correlation of the errors.
This is called three-stage least squares (3SLS).
Formally, the  that minimizes
that minimizes

is the 3SLS estimator of the parameters. For more details on
3SLS, refer to Gallant (1987, p. 435).
Residuals from the 2SLS method are used to estimate the  matrix
required for 3SLS.
The results of the preliminary 2SLS step are not printed unless the
2SLS option is also specified.
matrix
required for 3SLS.
The results of the preliminary 2SLS step are not printed unless the
2SLS option is also specified.
To use the three-stage least-squares method,
specify an INSTRUMENTS statement
and use the 3SLS or N3SLS option on either the PROC MODEL statement
or a FIT statement.
Generalized Method of Moments - GMM
For systems of equations with heteroscedastic errors, generalized
method of moments (GMM) can be used to obtain
efficient estimates of the parameters.
See the "Heteroscedasticity" section
for alternatives to GMM.
Consider the nonlinear model

where zt is a vector of instruments and
 t is an unobservable disturbance
vector that can be serially correlated and nonstationary.
t is an unobservable disturbance
vector that can be serially correlated and nonstationary.
In general, the following orthogonality condition
is desired:

which states that the expected crossproducts of the unobservable
disturbances,  , and functions of the
observable variables are set to 0. The first moment of the
crossproducts is
, and functions of the
observable variables are set to 0. The first moment of the
crossproducts is

where 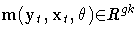 .
.
The case where gk > p is considered here, where p is
the number of parameters.
Estimate the true parameter vector  by the value of
by the value of 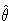 that minimizes
that minimizes
![S({\theta}, V) = [n{m}_{n}({\theta})]' V\hspace*{1pt}^{-1}[n{m}_{n}({\theta})]](images/modeq80.gif)
where
![V = \rm{Cov}([n{m}_{n}({\theta}^0)], [n{m}_{n}({\theta}^0)]')](images/modeq81.gif)
The parameter vector that minimizes this objective function
is the GMM estimator.
GMM estimation is requested
on the FIT statement with the GMM option.
The variance of the moment functions, V, can be
expressed as
![V
&=&
E (\sum_{t=1}^n{{{\epsilon}}_{t} {\otimes} z_{t}})
(\sum_{s=1}^n{{{\eps...
...} {\otimes} z_{t})
( {{\epsilon}}_{s} {\otimes} z_{s})']}
\cr
&=&
n S_{n}^0](images/modeq82.gif)
where Sn0 is estimated as

Note that  is a gk×gk matrix.
Because Var
is a gk×gk matrix.
Because Var 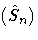 will not decrease with
increasing n
we consider estimators of Sn0 of the
form:
will not decrease with
increasing n
we consider estimators of Sn0 of the
form:
![\hat{S}_{n}(l(n)) &=&
\sum_{{\tau} = -n + 1}^{n-1}{w {{\tau} \overwithdelims ()...
...t-{\tau}}]'} &
{\tau}\gt=0\space \cr
(\hat{S}_{n,-{\tau}})' &
{\tau}\lt\cr
}](images/modeq86.gif)
where l(n) is a scalar function that computes the bandwidth parameter,
w(·) is a scalar
valued kernel, and the diagonal matrix D is used for a
small sample degrees of freedom correction (Gallant 1987).
The initial  used for the estimation of
used for the estimation of  is obtained
from a 2SLS estimation of the system.
The degrees of freedom correction is handled by the
VARDEF= option as for the S matrix estimation.
is obtained
from a 2SLS estimation of the system.
The degrees of freedom correction is handled by the
VARDEF= option as for the S matrix estimation.
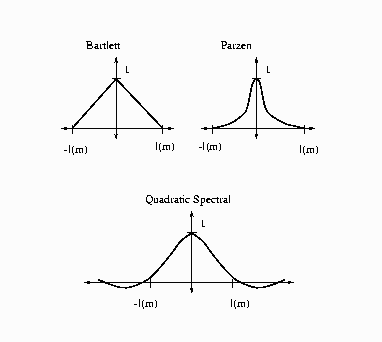
The following kernels are supported by PROC MODEL. They are listed
with their default bandwidth functions:
Bartlett: KERNEL=BART

Parzen: KERNEL=PARZEN

Quadratic Spectral: KERNEL=QS

Figure 14.15: Kernels for Smoothing
Details of the properties of these and other kernels are given in
Andrews (1991).
Kernels are selected with the KERNEL= option; KERNEL=PARZEN is
the default. The general form of the KERNEL= option is
KERNEL=( PARZEN | QS | BART, c, e )
where the e >= 0 and c >= 0 are used to compute the bandwidth
parameter as
-
l(n) = c ne
The bias of the standard error estimates increases for
large bandwidth parameters. A warning message is produced for
bandwidth parameters greater than n(1/3).
For a discussion
of the computation of the optimal l(n), refer to Andrews (1991).
The "Newey-West" kernel (Newey (1987)) corresponds to the Bartlett
kernel with bandwith parameter l(n) = L +1. That is, if the
"lag length" for the Newey-West kernel is L then the
corresponding Model procedure syntax is KERNEL=( bart, L+1, 0).
Andrews (1992) has shown that using prewhitening in combination with
GMM can improve
confidence interval coverage and reduce over rejection of t-statistics
at the cost of inflating the variance and MSE of the estimator. Prewhitening
can be performed using the %AR macros.
For the special case that the errors are not serially correlated, that is

the estimate for Sn0 reduces to
![\hat{S}_{n} = \frac{1}n
\sum_{t=1}^n{[q(y_{t}\hspace*{1pt},
x_{t}\hspace*{1pt}...
..._{t}] [q(y_{t}\hspace*{1pt}, x_{t}\hspace*{1pt},
{{\theta}}) {\otimes} z_{t}]'}](images/modeq92.gif)
The option KERNEL=(kernel,0,) is used to select this type of
estimation when using GMM.
Testing Over-Identifying Restrictions
Let r be the number of unique instruments times the number of equations.
The value r represents the number of orthogonality conditions imposed
by the GMM method.
Under the assumptions of the GMM method,
r-p linearly independent combinations of the orthogonality
should be close to zero. The GMM estimates are computed by setting
these combinations to zero.
When r exceeds the number of parameters to be estimated,
the OBJECTIVE*N, reported at the end of the estimation, is an asymptoticly
valid statistic to test the null hypothesis that the over-identifying
restrictions of the model are valid. The OBJECTIVE*N is distributed
as a chi-square with r-p degrees of freedom (Hansen 1982, p. 1049).
Iterated Generalized Method of Moments - ITGMM
Iterated generalized method of moments is similar to the
iterated versions of 2SLS, SUR, and 3SLS. The variance matrix for
GMM estimation
is re-estimatedg at each iteration with the parameters determined by
the GMM estimation. The iteration terminates when the variance matrix
for the equation errors change less than the CONVERGE= value. Iterated
generalized method of moments is selected by the ITGMM option on the
FIT statement. For some indication of the small sample properties of
ITGMM, refer to Ferson (1993).
Full Information Maximum Likelihood Estimation - FIML
A different approach to the simultaneous equation bias problem
is the full information maximum likelihood (FIML) estimation method
(Amemiya 1977).
Compared to the instrumental variables methods (2SLS and 3SLS),
the FIML method has these advantages and disadvantages:
- FIML does not require instrumental variables.
- FIML requires that the model include the full equation system,
with as many equations as there are endogenous variables.
With 2SLS or 3SLS you can estimate some of the equations
without specifying the complete system.
- FIML assumes that the equations errors have a multivariate
normal distribution. If the errors are not normally distributed,
the FIML method may produce poor results.
2SLS and 3SLS do not assume a specific distribution for the errors.
- The FIML method is computationally expensive.
The full information maximum likelihood estimators of  and
and
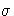 are
the
are
the 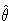 and
and 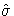 that minimize
the negative log likelihood function:
that minimize
the negative log likelihood function:

The option FIML requests full information maximum likelihood estimation.
If the errors are distributed normally, FIML produces efficient estimators
of the parameters. If instrumental variables are not provided the
starting values for the estimation are obtained from a SUR estimation.
If instrumental variables are provided, then the starting
values are obtained from a 3SLS estimation. The negative log likelihood value
and the l2 norm of the gradient of the negative log likelihood function
are shown in the estimation summary.
FIML Details
To compute the minimum of  ,this function is concentrated using the relation:
,this function is concentrated using the relation:

This results in the concentrated negative log likelihood function:

The gradient of the negative log likelihood function is :


where

The estimator of the variance-covariance of 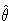 (COVB)
for FIML can be selected with the COVBEST= option with the following arguments:
(COVB)
for FIML can be selected with the COVBEST= option with the following arguments:
- CROSS
- selects the crossproducts estimator of the covariance matrix (default)
(Gallant 1987, p. 473):

where ![{{\nabla}(t) = [{\nabla}_{1}(t), {\nabla}_{2}(t), { ... },{\nabla}_{p}(t)]'}](images/modeq103.gif)
- GLS
- selects the generalized least-squares estimator
of the covariance matrix. This is computed as (Dagenais 1978)
![C = [ \hat{Z} ' ( {\Sigma}({\theta})^{-1} {\otimes} I)
\hat{Z}]^{-1}](images/modeq104.gif)
where  is ng ×p and each
is ng ×p and each 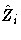 column vector is
obtained from stacking the columns of
column vector is
obtained from stacking the columns of

U is an n ×g matrix of residuals and qi
is an n ×g matrix
 .
.
- FDA
- selects the inverse of concentrated likelihood Hessian
as an estimator of the covariance matrix. The Hessian is computed
numerically, so for a large problem this is computationally expensive.
The HESSIAN= option controls which approximation to the Hessian is
used in the minimization procedure. Alternate approximations
are used to improve convergence and execution time. The choices are
- CROSS
- The crossproducts approximation is used.
- GLS
- The generalized least-squares approximation is used (default).
- FDA
- The Hessian is computed numerically by finite differences.
HESSIAN=GLS has better convergence properties in general,
but COVBEST=CROSS produces the most pessimistic standard error bounds.
When the HESSIAN= option is used, the default estimator of the
variance-covariance of 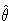 is the inverse of
the Hessian selected.
is the inverse of
the Hessian selected.
Properties of the Estimates
All of the methods are consistent.
Small sample properties may not be good for nonlinear models.
The tests and standard errors
reported are based on the convergence of the distribution of the
estimates to a normal distribution in large samples.
These nonlinear estimation methods reduce to the corresponding linear
systems regression methods if the model is linear.
If this is the case, PROC MODEL produces the same estimates as PROC SYSLIN.
Except for GMM, the estimation methods assume that the equation errors
for each observation are
identically and independently distributed with a 0 mean vector and
positive definite covariance matrix  consistently estimated by
S. For FIML, the errors need to be normally distributed.
There are no other assumptions concerning the distribution of
the errors for the other estimation methods.
consistently estimated by
S. For FIML, the errors need to be normally distributed.
There are no other assumptions concerning the distribution of
the errors for the other estimation methods.
The consistency of the parameter estimates relies on the assumption
that the S matrix is a consistent estimate of  .These standard error estimates are asymptotically valid, but for nonlinear
models they may not be reliable for small samples.
.These standard error estimates are asymptotically valid, but for nonlinear
models they may not be reliable for small samples.
The S matrix used for the calculation of the covariance of the parameter
estimates is the best estimate available
for the estimation method selected. For S-iterated methods this
is the most recent estimation of  . For OLS and 2SLS,
an estimate of the S matrix is computed from OLS or 2SLS residuals and
used for the calculation of the covariance matrix. For a complete
list of the S matrix used for the calculation of the covariance of
the parameter estimates, see Table 14.1.
. For OLS and 2SLS,
an estimate of the S matrix is computed from OLS or 2SLS residuals and
used for the calculation of the covariance matrix. For a complete
list of the S matrix used for the calculation of the covariance of
the parameter estimates, see Table 14.1.
Missing Values
An observation is excluded from the estimation if any variable used
for FIT tasks is missing,
if the weight for the observation is not greater
than 0 when weights are used, or if a DELETE statement is executed by
the model program. Variables used for FIT tasks include the
equation errors for each equation, the instruments, if any, and the
derivatives of the equation errors with respect to the parameters
estimated. Note that variables can become missing as a result of
computational errors or calculations with missing values.
The number of usable observations can change when different parameter
values are used; some parameter values can be invalid and cause
execution errors for some observations. PROC MODEL keeps track of the
number of usable and missing observations at each pass through the data,
and if the number of missing observations counted during a pass exceeds
the number that was obtained using the previous parameter vector, the
pass is terminated and the new parameter vector is considered infeasible.
PROC MODEL never takes a step that produces more missing observations than
the current estimate does.
The values used to compute the Durbin-Watson, R2,
and other statistics of fit are from the observations used
in calculating the objective function and do not include any
observation for which any needed variable was missing
(residuals, derivatives, and instruments).
Details on the Covariance of Equation Errors
There are several S matrices that can be involved in the various
estimation methods and in forming the estimate of the covariance of
parameter estimates. These S matrices are estimates of  ,the true covariance of the equation errors.
Apart from the choice of instrumental or noninstrumental methods,
many of the methods provided by PROC MODEL differ
in the way the various S matrices are formed and used.
,the true covariance of the equation errors.
Apart from the choice of instrumental or noninstrumental methods,
many of the methods provided by PROC MODEL differ
in the way the various S matrices are formed and used.
All of the estimation methods result in a final estimate of  ,which is included in the output if the COVS
option is specified. The final S matrix of each method provides the
initial S matrix for any subsequent estimation.
,which is included in the output if the COVS
option is specified. The final S matrix of each method provides the
initial S matrix for any subsequent estimation.
This estimate of the covariance of equation errors is defined as
-
S = D(R'R)D
where R = (r1, ... ,rg)
is composed of the equation residuals computed from the current parameter
estimates in an n ×g matrix and D is a diagonal matrix
that depends on the VARDEF= option.
For VARDEF=N, the diagonal elements of D are 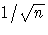 ,where n is the number of nonmissing observations.
For VARDEF=WGT, n is replaced with the sum of the weights.
For VARDEF=WDF, n is replaced with the sum of the weights minus
the model degrees of freedom.
For the default VARDEF=DF, the ith diagonal element of D is
,where n is the number of nonmissing observations.
For VARDEF=WGT, n is replaced with the sum of the weights.
For VARDEF=WDF, n is replaced with the sum of the weights minus
the model degrees of freedom.
For the default VARDEF=DF, the ith diagonal element of D is
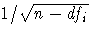 , where dfi is
the degrees of freedom (number of parameters) for the ith
equation. Binkley and Nelson (1984) show the importance of using a
degrees-of-freedom correction in estimating
, where dfi is
the degrees of freedom (number of parameters) for the ith
equation. Binkley and Nelson (1984) show the importance of using a
degrees-of-freedom correction in estimating  . Their
results indicate that the DF method produces more
accurate confidence intervals for N3SLS parameter estimates in the
linear case than the alternative approach they tested. VARDEF=N
is always used for the computation of the FIML estimates.
. Their
results indicate that the DF method produces more
accurate confidence intervals for N3SLS parameter estimates in the
linear case than the alternative approach they tested. VARDEF=N
is always used for the computation of the FIML estimates.
For the fixed S methods, the OUTSUSED= option writes
the S matrix used in the estimation to a data set. This S matrix
is either the estimate of
the covariance of equation errors matrix from the preceding estimation,
or a prior  estimate read in from a data set
when the SDATA= option is specified.
For the diagonal S methods, all of the off-diagonal elements of the S matrix
are set to 0 for the estimation of the parameters and for the OUTSUSED=
data set, but the output data set produced by
the OUTS= option will contain the off-diagonal elements.
For the OLS and N2SLS methods, there is no previous estimate of the
covariance of equation errors matrix, and the option OUTSUSED=
will save an identity matrix
unless a prior
estimate read in from a data set
when the SDATA= option is specified.
For the diagonal S methods, all of the off-diagonal elements of the S matrix
are set to 0 for the estimation of the parameters and for the OUTSUSED=
data set, but the output data set produced by
the OUTS= option will contain the off-diagonal elements.
For the OLS and N2SLS methods, there is no previous estimate of the
covariance of equation errors matrix, and the option OUTSUSED=
will save an identity matrix
unless a prior  estimate is supplied by the SDATA= option.
For FIML the OUTSUSED= data set contains the S matrix computed
with VARDEF=N. The OUTS= data set contains the S matrix computed
with the selected VARDEF= option.
If the COVS option is used, the method is not S-iterated,
and S is not an identity, the OUTSUSED= matrix is included
in the printed output.
estimate is supplied by the SDATA= option.
For FIML the OUTSUSED= data set contains the S matrix computed
with VARDEF=N. The OUTS= data set contains the S matrix computed
with the selected VARDEF= option.
If the COVS option is used, the method is not S-iterated,
and S is not an identity, the OUTSUSED= matrix is included
in the printed output.
For the methods that iterate the covariance of equation errors matrix,
the S matrix is iteratively re-estimated from the residuals produced by the
current parameter estimates.
This S matrix estimate iteratively replaces the previous estimate until
both the parameter estimates and the estimate of the covariance
of equation errors matrix converge.
The final OUTS= matrix and OUTSUSED= matrix are thus identical
for the S-iterated methods.
Nested Iterations
By default, for S-iterated methods, the S matrix is held constant until the
parameters converge once. Then the S matrix is re-estimated. One
iteration of the parameter estimation algorithm is performed, and
the S matrix is again re-estimated. This latter process is repeated
until convergence of both the parameters and the S matrix.
Since the objective of the
minimization depends on the S matrix, this has the effect of
chasing a moving target.
When the NESTIT option is specified, iterations are performed to
convergence for the structural parameters with a fixed S matrix.
The S matrix is then re-estimated, the parameter iterations
are repeated to convergence,
and so on until both the parameters and the S matrix
converge. This has the effect of fixing the objective function for
the inner parameter iterations.
It is more reliable, but usually more expensive, to nest the iterations.
R2
For unrestricted linear models with an intercept successfully
estimated by OLS, R2 is always between 0 and 1.
However, nonlinear models do not necessarily encompass the dependent mean
as a special case and can produce negative R2 statistics.
Negative R2's can also be produced even for linear models when
an estimation method other than OLS is used and no intercept term
is in the model.
R2 is defined for normalized equations as

where SSA is the sum of the squares of the actual y's
and 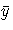 are the actual means.
R2 cannot be computed for models in general form because of
the need for an actual Y.
are the actual means.
R2 cannot be computed for models in general form because of
the need for an actual Y.
Copyright © 1999 by SAS Institute Inc., Cary, NC, USA. All rights reserved.


![{{r } = [\matrix{r_{1} \cr r_{2} \cr {\vdots} \cr r_{g}}] }](images/modeq48.gif) is the ng ×1 vector of residuals for the g equations stacked together.
is the ng ×1 vector of residuals for the g equations stacked together.
![{r_{i} = [\matrix{q_{i}(y_{1}\hspace*{1pt}, x_{1}\hspace*{1pt}, {{\theta}}) \cr ...
...) \cr {\vdots} \cr q_{i}(y_{n}\hspace*{1pt}, x_{n}\hspace*{1pt}, {{\theta}}) }]}](images/modeq49.gif) is the n ×1 column vector of residuals for the ith equation.
is the n ×1 column vector of residuals for the ith equation.











![S({\theta}, V) = [n{m}_{n}({\theta})]' V\hspace*{1pt}^{-1}[n{m}_{n}({\theta})]](images/modeq80.gif)
![V = \rm{Cov}([n{m}_{n}({\theta}^0)], [n{m}_{n}({\theta}^0)]')](images/modeq81.gif)
![V
&=&
E (\sum_{t=1}^n{{{\epsilon}}_{t} {\otimes} z_{t}})
(\sum_{s=1}^n{{{\eps...
...} {\otimes} z_{t})
( {{\epsilon}}_{s} {\otimes} z_{s})']}
\cr
&=&
n S_{n}^0](images/modeq82.gif)

![\hat{S}_{n}(l(n)) &=&
\sum_{{\tau} = -n + 1}^{n-1}{w {{\tau} \overwithdelims ()...
...t-{\tau}}]'} &
{\tau}\gt=0\space \cr
(\hat{S}_{n,-{\tau}})' &
{\tau}\lt\cr
}](images/modeq86.gif)




![\hat{S}_{n} = \frac{1}n
\sum_{t=1}^n{[q(y_{t}\hspace*{1pt},
x_{t}\hspace*{1pt}...
..._{t}] [q(y_{t}\hspace*{1pt}, x_{t}\hspace*{1pt},
{{\theta}}) {\otimes} z_{t}]'}](images/modeq92.gif)







![C = [ \hat{Z} ' ( {\Sigma}({\theta})^{-1} {\otimes} I)
\hat{Z}]^{-1}](images/modeq104.gif)

