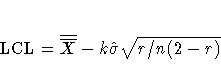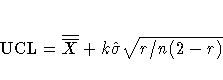Dictionary of Special Options
- ALPHA=value
-
requests probability limits.
If you specify ALPHA=
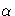 , the control limits are computed
so that the probability is
, the control limits are computed
so that the probability is 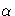 that a single EWMA
exceeds its control limits.
The value of
that a single EWMA
exceeds its control limits.
The value of 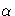 can range between 0 and 1.
This assumes that the
process is in statistical control and that the data follow a
normal distribution. For the equations used to compute probability
limits, see "Control Limits" .
can range between 0 and 1.
This assumes that the
process is in statistical control and that the data follow a
normal distribution. For the equations used to compute probability
limits, see "Control Limits" .
Note the following:
-
As an alternative to specifying ALPHA=
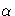 , you can read
, you can read
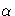 from the variable _ALPHA_ in a LIMITS= data set by
specifying the READALPHA option.
from the variable _ALPHA_ in a LIMITS= data set by
specifying the READALPHA option.
-
As an alternative to specifying ALPHA=
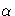 (or reading _ALPHA_ from a LIMITS= data set),
you can request "
(or reading _ALPHA_ from a LIMITS= data set),
you can request "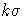 control limits" by
specifying SIGMAS=k
(or reading _SIGMAS_ from a LIMITS= data set).
control limits" by
specifying SIGMAS=k
(or reading _SIGMAS_ from a LIMITS= data set).
If you specify neither the ALPHA= option nor the SIGMAS= option, the
procedure computes 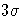 control limits by default.
control limits by default.
- ASYMPTOTIC
-
requests constant upper and lower control limits based on the following
asymptotic expressions:
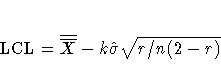
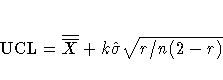
Here r is the weight parameter 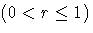 , and n is
the nominal sample size associated with the control limits.
Substitute
, and n is
the nominal sample size associated with the control limits.
Substitute 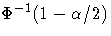 for k if you specify
probability limits with the ALPHA= option.
When you do not specify the ASYMPTOTIC option,
the control limits are computed using the exact formulas
in Table 20.19.
Use the ASYMPTOTIC option only if all the subgroup sample
sizes are the same or if you specify LIMITN=n.
See Example 20.2.
for k if you specify
probability limits with the ALPHA= option.
When you do not specify the ASYMPTOTIC option,
the control limits are computed using the exact formulas
in Table 20.19.
Use the ASYMPTOTIC option only if all the subgroup sample
sizes are the same or if you specify LIMITN=n.
See Example 20.2.
- CMEANSYMBOL=color
-
[Graphics]
specifies the color for the symbol requested with the
MEANSYMBOL= option. The default color is the first
color in the device color list.
- LIMITN=n
- LIMITN=VARYING
-
specifies either a fixed or varying nominal sample
size for the control limits.
If you specify LIMITN=n,
EWMAs are calculated and displayed only for those
subgroups with a sample size
equal to n, unless you also specify the ALLN option,
which causes all
the EWMAs to be calculated and displayed.
By default (or if you specify LIMITN=VARYING),
EWMAs are calculated and displayed for all subgroups, regardless of
sample size.
- MEANCHAR='character'
-
[Line Printer]
specifies a character used to plot the subgroup mean for each subgroup.
By default, subgroup means are not plotted.
- MEANSYMBOL=keyword
-
[Graphics]
specifies a symbol used to plot the subgroup mean for each subgroup.
By default, subgroup means are not plotted.
- MU0=value
-
specifies a known (standard) value
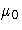 for the process
mean
for the process
mean 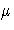 . By default,
. By default, 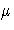 is estimated from the data.
See Example 20.1.
is estimated from the data.
See Example 20.1.
Note:
As an alternative to specifying MU0=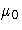 , you can read a
predetermined value for
, you can read a
predetermined value for 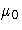 from the variable _MEAN_
in a LIMITS= data set.
from the variable _MEAN_
in a LIMITS= data set.
- NOREADLIMITS
-
specifies that control limit parameters for each process
listed in the EWMACHART statement are not to be read from the
LIMITS= data set specified in the PROC MACONTROL statement. The
NOREADLIMITS option is available only in Release 6.10 and later
releases.
The following example illustrates the NOREADLIMITS option:
proc macontrol data=pistons limits=diamlim;
ewmachart diameter*hour;
ewmachart diameter*hour / noreadlimits weight=0.3;
run;
The first EWMACHART statement reads the control limits from
the first observation in the data set DIAMLIM for which
the variable _VAR_ is equal to diameter and the
variable _SUBGRP_ is equal to hour. The second
EWMACHART statement computes estimates of the process
mean and standard deviation for the control limits from the
measurements in the data set PISTONS. Note that the
second EWMACHART statement is equivalent to the following
statements, which would be more commonly used:
proc macontrol data=pistons;
ewmachart diameter*hour / weight=0.3;
run;
For more information about reading control limit parameters from
a LIMITS= data set, see the READLIMITS
option later in this list.
- READALPHA
-
specifies that the variable _ALPHA_, rather than the variable
_SIGMAS_, is to be read from a LIMITS= data set when both
variables are available in the data set.
Thus the limits displayed are probability limits.
If you do not specify the READALPHA option, then _SIGMAS_ is
read by default.
- READINDEX='value'
-
reads control limit parameters from a LIMITS= data set
(specified in the PROC MACONTROL statement) for each process
listed in the EWMACHART statement.
The control limit parameters
for a particular process are read from the
first observation in the LIMITS= data set for which
- the value of _VAR_ matches process
- the value of _SUBGRP_ matches the subgroup-variable
- the value of _INDEX_ matches value
The value can be up to 16 characters and must be enclosed
in quotes.
- READLIMITS
-
specifies that control limit parameters are to be read from a
LIMITS= data set specified in the PROC MACONTROL statement.
The parameters for a particular process are read from the
first observation in the LIMITS= data set for which
- the value of _VAR_ matches process
- the value of _SUBGRP_ matches the subgroup variable
The use of the READLIMITS option depends on which release
of SAS/QC software you are using.
- In Release 6.10 and later releases, the READLIMITS
option is not necessary. To read control limits parameters as
described previously, you simply specify a LIMITS= data set.
However, even though the READLIMITS option is redundant,
it continues to function as in earlier releases.
- In Release 6.09 and earlier releases, you must
specify the READLIMITS option to read control limits parameters
as described previously. If you specify a LIMITS= data
set without specifying the READLIMITS option (or the
READINDEX= option), the control limits are computed from the
data and the value of the weight parameter is specified
with the WEIGHT= option.
- RESET
-
requests that the value of the EWMA be reset
after each out-of-control point. Specifically, when a point
exceeds the control limits, the EWMA for the next subgroup
is computed as the weighted average of the subgroup mean and
the overall mean.
By default, the EWMAs are not reset.
- SIGMA0=value
-
specifies a known (standard) value
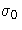 for the
process standard deviation
for the
process standard deviation 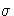 . The value
must be positive. By default, the MACONTROL procedure estimates
. The value
must be positive. By default, the MACONTROL procedure estimates
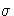 from the data using the formulas given in
"Methods for Estimating the Standard Deviation"
.
from the data using the formulas given in
"Methods for Estimating the Standard Deviation"
.
Note:
As an alternative to specifying SIGMA0=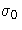 ,you can read a predetermined value for
,you can read a predetermined value for 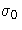 from the variable _STDDEV_ in a LIMITS= data set.
from the variable _STDDEV_ in a LIMITS= data set.
- SIGMAS=value
-
specifies the width of the control limits in terms of the
multiple k of the standard error of the plotted EWMAs
on the chart. The value of k must be
positive. By default, k=3 and the control limits are
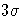 limits.
limits.
- WEIGHT=value
-
specifies the weight r assigned to the most recent subgroup mean in
the computation of the EWMA
(
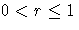 ). The WEIGHT= option is required unless
you read control limit parameters from a LIMITS= data set or a
TABLE= data set. See "Choosing the Value of the Weight Parameter"
for
details.
). The WEIGHT= option is required unless
you read control limit parameters from a LIMITS= data set or a
TABLE= data set. See "Choosing the Value of the Weight Parameter"
for
details.
Copyright © 1999 by SAS Institute Inc., Cary, NC, USA. All rights reserved.