STAT 350: Lecture 34
Non-linear Regression
In a regression model of the form
![]()
we call the model linear if ![]() for some covariates
for some covariates
![]() and a parameter
and a parameter ![]() . Often, however, we have non-linear
models such as
. Often, however, we have non-linear
models such as ![]() or in general
or in general ![]() for some function f which need not
be linear in
for some function f which need not
be linear in ![]() . If the errors are
homo-scedastic normal errors then the maximum likelihood estimates of
the
. If the errors are
homo-scedastic normal errors then the maximum likelihood estimates of
the ![]() are least squares estimates, minimizing
are least squares estimates, minimizing
![]() . The value
. The value ![]() then solves
then solves
![]()
This equation can usually not be solved analytically. In general the process of solving it numerically is iterative. One approach is:
![]()
where ![]() is the vector of partial derivatives of
is the vector of partial derivatives of ![]() with respect to
the entries in
with respect to
the entries in ![]() .
.
![]()
with respect to ![]() .
.
![]()
where ![]() . The solution is
. The solution is
![]() . We compute this
. We compute this ![]() .
.
Example
In thermoluminescence dating ( see Lecture 1) a common model is
![]()
Thus in this case
![]()
The derivatives of ![]() with respect to the entries of
with respect to the entries of ![]() are
are
![]()
![]()
and
![]()
Software and computing details
In SAS the procedure proc nlin will do non-linear least squares.
You need to specify quite a few ingredients. The model statement must
specify the exact functional form of the function ![]() . You have a
statement parms which is needed to specify the initial guess for
. You have a
statement parms which is needed to specify the initial guess for ![]() .
You usually will have a bunch of der statements to specify derivatives of
the predicted values with respect to each parameter and, if you use some algorithms,
you will even specify some second derivatives. (You get a choice of 5 different
iterative algorithms. The one described above is called Gauss-Newton.)
.
You usually will have a bunch of der statements to specify derivatives of
the predicted values with respect to each parameter and, if you use some algorithms,
you will even specify some second derivatives. (You get a choice of 5 different
iterative algorithms. The one described above is called Gauss-Newton.)
In Splus the function nls can be used to to the same thing.
Here is a plot of some thermoluminescence data.
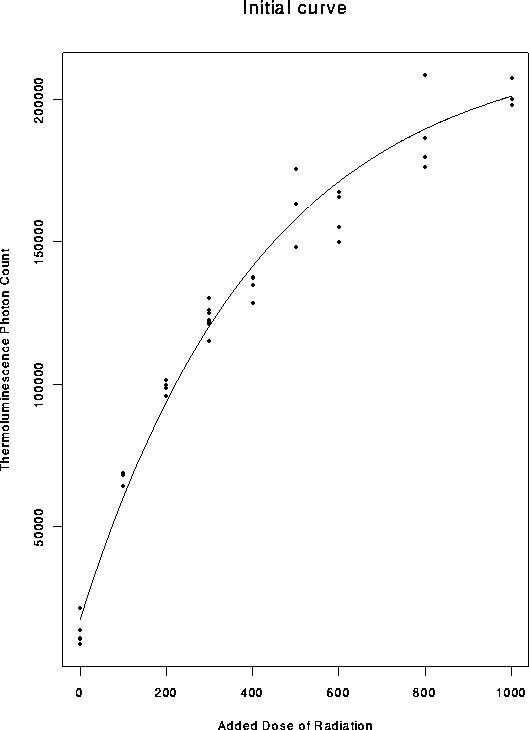
I will analyze the data by using the Gauss-Newton algorithm by hand and then using glm and nlin
S-PLUS : Copyright (c) 1988, 1996 MathSoft, Inc.
S : Copyright AT&T.
Version 3.4 Release 1 for Sun SPARC, SunOS 5.3 : 1996
Working data will be in .Data
> > dat <- read.table("satur.data",header=T)
> #
> # Read in data
> #
> beta0 <- 220000
> #
> # From looking at picture guess saturation level
> #
> beta <- lsfit(dat$Dose,-log(1-dat$Count/beta0))$coef
> #
> # Get initial values for beta by solving
> # y = b1(1-exp(-(x-b2)/b3)) to get
> # log(1-y/b1) = -(x-b2)/b3 and then regressing
> # log(1-y/b1) on dose.
> #
> beta <- lsfit(dat$Dose,-log(1-dat$Count/beta0))$coef
> beta[2] <- 1/beta[2]
> beta[1] <- -beta[2]*beta[1]
> beta <- c(beta0,beta)
> beta
Intercept X
220000 -34.46809 422.0629
> #
> # These are the initial estimates
> #
> dv <- seq(0,max(dat$Dose),length=300)
> cv <- beta[1]*(1-exp(-(dv-beta[2])/beta[3]))
> postscript("curve0.ps",horizontal=F)
> plot(dat$Dose, dat$Count,xlab="Added Dose of Radiation",
+ ylab="Thermoluminescence Photon Count",main="Initial curve")
> lines(dv,cv)
> dev.off()
Generated postscript file "curve0.ps".
null device
1
> #
> # Plot the initial curve on top of the data.
> #
> fder <- function(d,b){
+ #
+ # This function computes derivatives of
+ # mu with respect to each component of
+ # beta and assembles them into a matrix
+ #
+ v <- exp(-(d-b[2])/b[3])
+ cbind(1-v,-v*b[1]/b[3],-b[1]*(d-b[2])*v/b[3]^2)
+ }
> mu_function(d,b){
+ #
+ # This function computes the vector mu
+ # of fitted values
+ #
+ b[1]*(1-exp(-(d-b[2])/b[3]))
+ }
> #
> # Plot the initial curveon top of the data.
> #
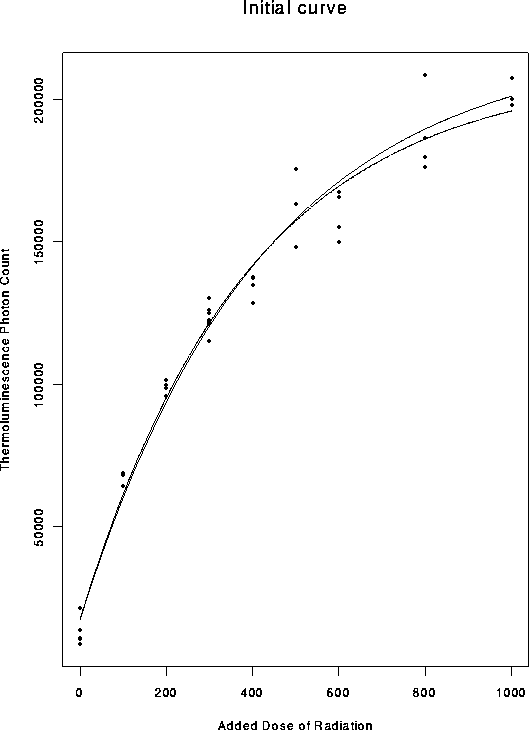
> postscript("curves.ps",horizontal=F)
> plot(dat$Dose, dat$Count,xlab="Added Dose of Radiation",
+ ylab="Thermoluminescence Photon Count",main="Initial curve")
> lines(dv,cv)
> #
> # Here we regress the original Ys minus the current fit on the
> # derivative of the fit with respect to the parameters.
> #
> ystar <- dat$Count -mu(dat$Dose,beta)
> delta <- lsfit(fder(dat$Dose,beta),ystar,intercept=F)$coef
> beta1 <- beta+delta
> beta <- beta1
> print(delta)
X1 X2 X3
-9433.193 0.8641427 -33.86906
> #
> # The increment in the fitted beta vector
> #
> cv <- beta[1]*(1-exp(-(dv-beta[2])/beta[3]))
> lines(dv,cv,lty=2)
> #
> # Add a curve to the plot for the new beta vector
> # and then repeat the process
> #
> ystar <- dat$Count -mu(dat$Dose,beta)
> delta <- lsfit( fder(dat$Dose,beta),ystar,intercept=F)$coef
> beta2 <- beta+delta
> print(delta)
X1 X2 X3
-115.2705 0.5834134 -1.532228
> beta <- beta2
> cv <- beta[1]*(1-exp(-(dv-beta[2])/beta[3]))
> lines(dv,cv,lty=3)
> #
> # Add a curve to the plot for the new beta vector
> # and then repeat the process
> #
> ystar <- dat$Count -mu(dat$Dose,beta)
> delta <- lsfit( fder(dat$Dose,beta),ystar,intercept=F)$coef
> beta3 <- beta+delta
> print(delta)
X1 X2 X3
-26.40946 0.02285527 -0.1350775
> beta <- beta3
> cv <- beta[1]*(1-exp(-(dv-beta[2])/beta[3]))
> lines(dv,cv,lty=4)
> #
> # Add a curve to the plot for the new beta vector
> # and then repeat the process
> #
> ystar <- dat$Count -mu(dat$Dose,beta)
> delta <- lsfit( fder(dat$Dose,beta),ystar,intercept=F)$coef
> beta4 <- beta+delta
> print(delta)
X1 X2 X3
-2.416176 0.002208632 -0.01256684
> beta <- beta4
> cv <- beta[1]*(1-exp(-(dv-beta[2])/beta[3]))
> lines(dv,cv,lty=5)
> #
> # Add a curve to the plot for the new beta vector
> # and then repeat the process
> #
# Intercept X
# beta0 220000.0 -34.46809 422.0629
# beta1 210566.8 -33.60395 388.1938
# beta2 210451.5 -33.02054 386.6616
# beta3 210425.1 -32.99768 386.5265
# beta4 210422.7 -32.99547 386.5139
> dev.off()
Generated postscript file "curves.ps".
null device
1
> q() # end-of-file
The successive fits are plotted here:
options pagesize=60 linesize=80; data thermo; infile 'satur.data' firstobs=2; input Code Dose Count ; proc nlin data=thermo; parms b1=220000 b2=-34.46809 b3=422.0629; model Count = b1*(1-exp(-(Dose - b2)/b3)); der.b1=(1-exp(-(Dose - b2)/b3)); der.b2=-b1*exp(-(Dose - b2)/b3)/b3; der.b3=-b1*(Dose - b2)*exp(-(Dose - b2)/b3)/b3**2; run;producing the output
Non-Linear Least Squares Iterative Phase
Dependent Variable COUNT Method: Gauss-Newton
Iter B1 B2 B3 Sum of Squares
0 220000 -34.468090 422.062900 2824928590
1 210567 -33.603952 388.193816 2670087246
2 210452 -33.020538 386.661589 2669527014
3 210425 -32.997683 386.526512 2669525464
4 210423 -32.995474 386.513945 2669525451
NOTE: Convergence criterion met.
Non-Linear Least Squares Summary Statistics Dependent Variable COUNT
Source DF Sum of Squares Mean Square
Regression 3 679415907744 226471969248
Residual 35 2669525451 76272156
Uncorrected Total 38 682085433194
(Corrected Total) 37 122311153163
Parameter Estimate Asymptotic Asymptotic 95 %
Std. Error Confidence Interval
Lower Upper
B1 210422.7105 6587.3455230 197049.76755 223795.65354
B2 -32.9955 8.2899354 -49.82484 -16.16611
B3 386.5139 31.3425426 322.88558 450.14231
Asymptotic Correlation Matrix
Corr B1 B2 B3
----------------------------------------------------------
B1 1 -0.442969052 0.9150075187
B2 -0.442969052 1 -0.663625494
B3 0.9150075187 -0.663625494 1