|
Kumar Abhishek
kabhisadasflkjhhe [at] sdfhsdjaffu
[dot] 1432@#$2 ca
I am a PhD student in the School of Computing Science at Simon Fraser University, where I work as a part of the Medical Image Analysis Lab (MIAL), under the supervision of
Professor Ghassan Hamarneh.
I defended my MSc Thesis in April 2020 on input space augmentation strategies for skin lesion
segmentation under the supervision of Professor Ghassan
Hamarneh. My examination committee consisted of Professors Mark S. Drew, Sandra Avila, and Angel X. Chang, and my thesis was accepted
without any revisions.
Previously, I graduated with a Bachelor of Technology in Electronics and Communication Engineering with
a focus on
Image Processing and Machine Learning from the Indian Institute of
Technology (IIT) Guwahati in 2015. My undergraduate thesis was advised by Professor Prithwijit Guha.
During my undergraduate years, I carried out internships at LFOVIA, IIT Hyderabad and CTO Office, Wipro. After graduating from IIT Guwahati, I have worked at Wipro Analytics and Altisource Labs.
Resume |
CV |
Google Scholar |
LinkedIn |
Blog
|

|
|
Research
I'm interested in computer vision, machine learning, and image processing. At MIAL, I work on applying
deep learning methods to medical image analysis. The primary focus of my work has been on skin lesion image analysis.
|
|
Analysis
Paper
|
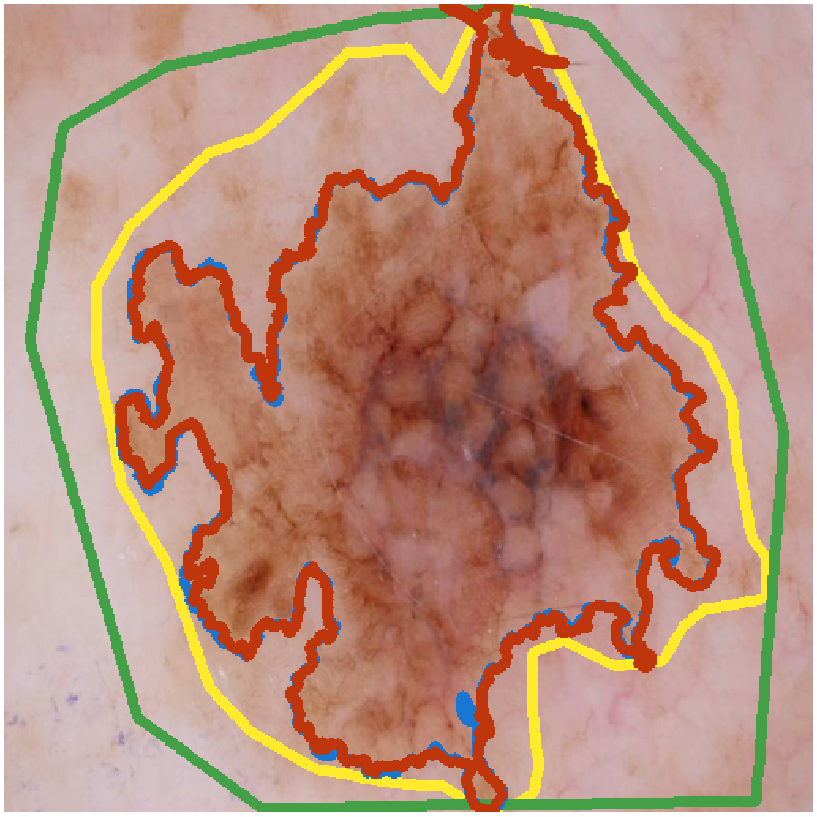
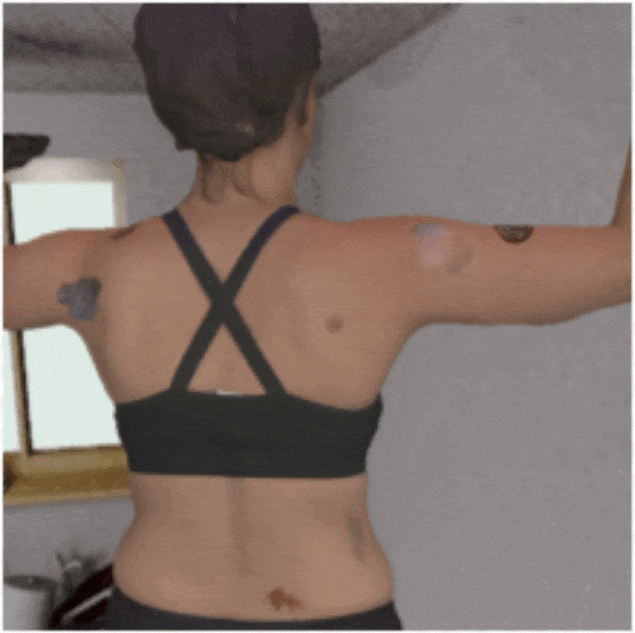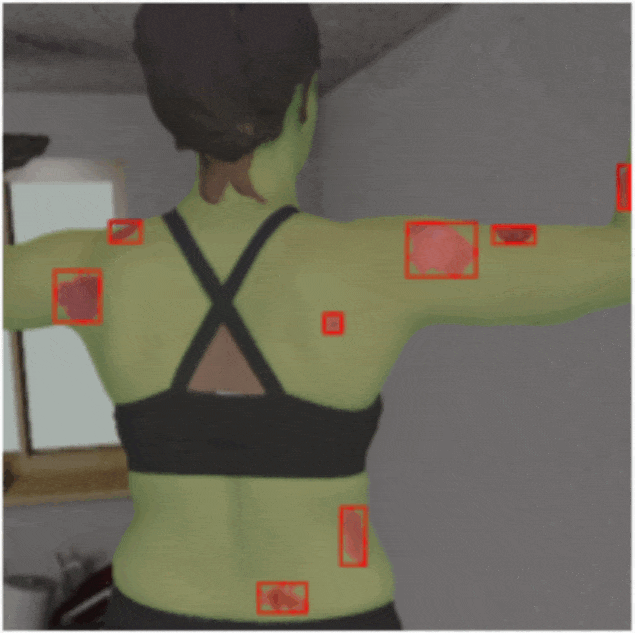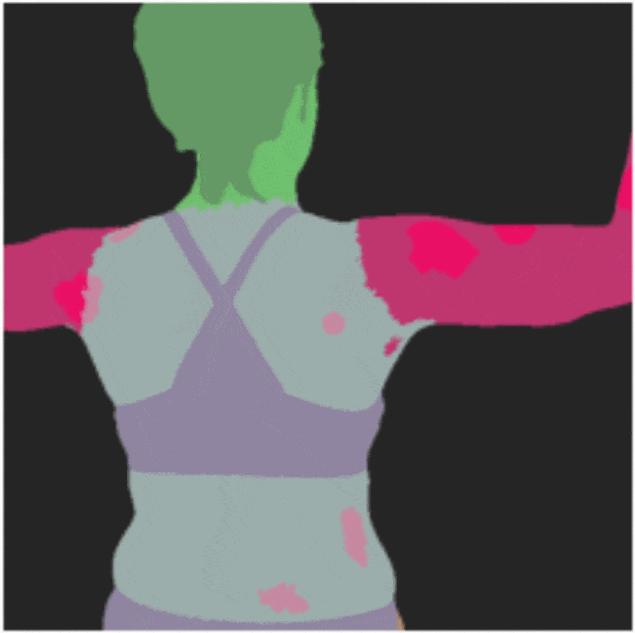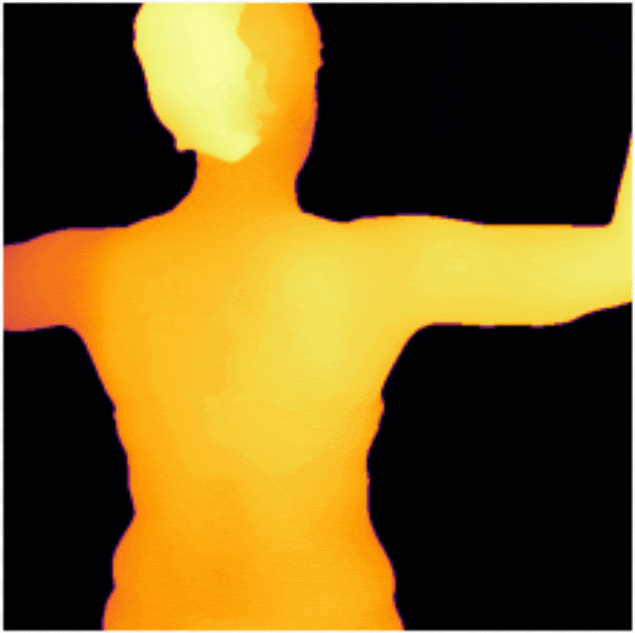
Investigating the Quality of DermaMNIST and Fitzpatrick17k Dermatological Image Datasets
Kumar Abhishek,
Aditi Jain,
Ghassan Hamarneh
under revision, 2024
We present an in-depth analysis of two popular dermatological image datasets, DermaMNIST
and Fitzpatrick17k, uncovering data quality issues: duplicates, data leakage across
train-test partitions, mislabeled images, and the absence of a well-defined test partition.
[Abstract]
The remarkable progress of deep learning in dermatological tasks has brought us closer
to achieving diagnostic accuracies comparable to those of human experts. However, while
large datasets play a crucial role in the development of reliable deep neural network
models, the quality of data therein and their correct usage are of paramount importance.
Several factors can impact data quality, such as the presence of duplicates, data
leakage across train-test partitions, mislabeled images, and the absence of a
well-defined test partition. In this paper, we conduct meticulous analyses of two
popular dermatological image datasets: DermaMNIST and Fitzpatrick17k, uncovering these
data quality issues, measure the effects of these problems on the benchmark results,
and propose corrections to the datasets. Besides ensuring the reproducibility of our
analysis, by making our analysis pipeline and the accompanying code publicly available,
we aim to encourage similar explorations and to facilitate the identification and
addressing of potential data quality issues in other large datasets.
|
|
DermSynth3D: Synthesis of in-the-wild Annotated Dermatology Images
Jeremy Kawahara,
Ashish Sinha,
Arezou Pakzad,
Kumar Abhishek,
Matthieu Ruthven,
Enjie Ghorbel,
Anis Kacem,
Djamila Aouada,
Ghassan Hamarneh
Medical Image Analysis,
2024
We propose a framework to synthesize in-the-wild 2D clinical images of skin diseases and provide
corresponding annotations for several downstream tasks.
[Abstract]
In recent years, deep learning (DL) has shown great potential in the field of dermatological
image analysis. However, existing datasets in this domain have significant limitations,
including a small number of image samples, limited disease conditions, insufficient annotations,
and non-standardized image acquisitions. To address these shortcomings, we propose a novel
framework called DermSynth3D. DermSynth3D blends skin disease patterns onto 3D textured meshes
of human subjects using a differentiable renderer and generates 2D images from various camera
viewpoints under chosen lighting conditions in diverse background scenes. Our method adheres to
top-down rules that constrain the blending and rendering process to create 2D images with skin
conditions that mimic in-the-wild acquisitions, ensuring more meaningful results. The framework
generates photo-realistic 2D dermoscopy images and the corresponding dense annotations for
semantic segmentation of the skin, skin conditions, body parts, bounding boxes around lesions,
depth maps, and other 3D scene parameters, such as camera position and lighting conditions.
DermSynth3D allows for the creation of custom datasets for various dermatology tasks. We
demonstrate the effectiveness of data generated using DermSynth3D by training DL models on
synthetic data and evaluating them on various dermatology tasks using real 2D dermatological
images. We make our code publicly available on GitHub.
|
|
|
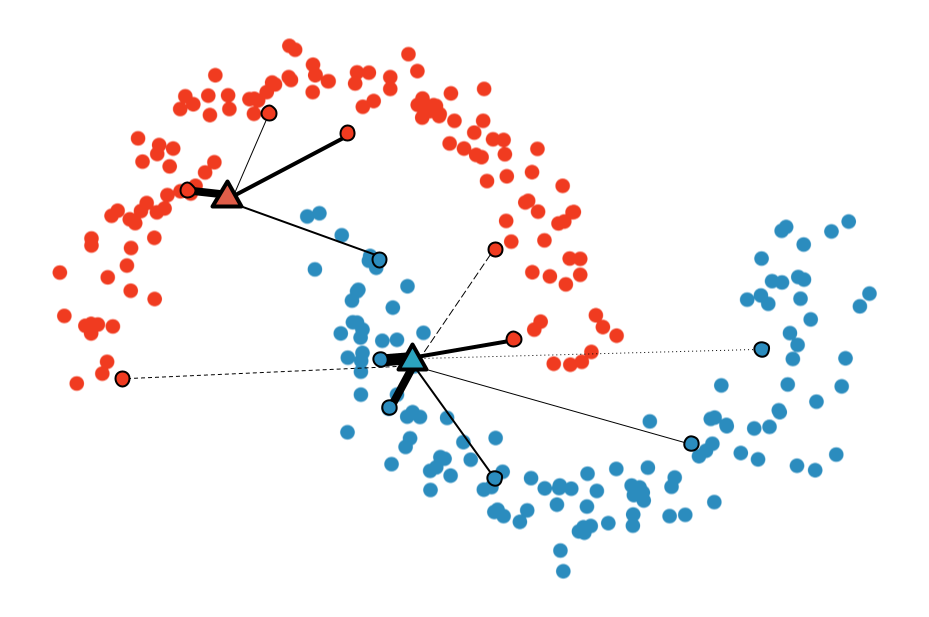
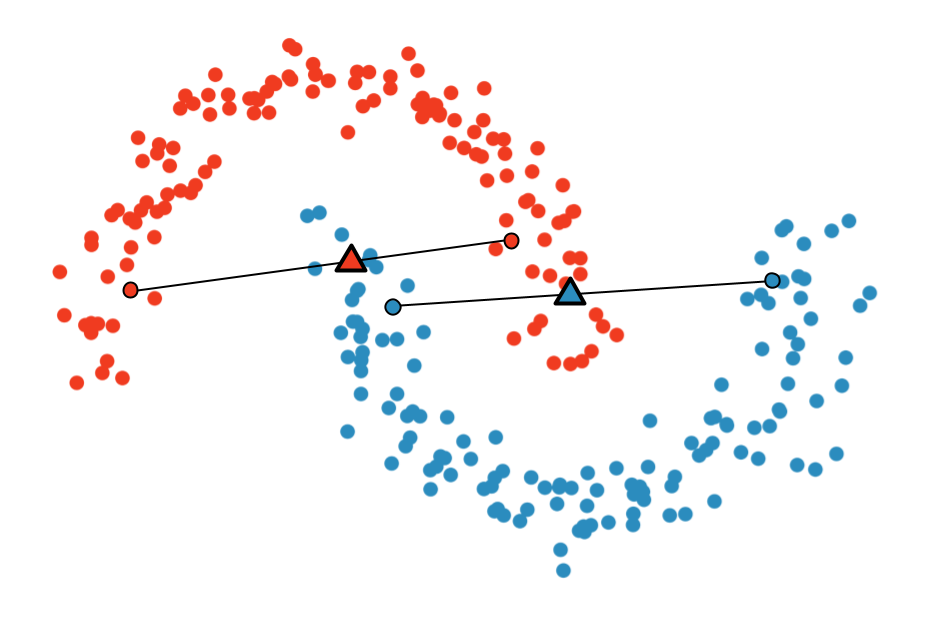
Multi-Sample ζ-mixup: Richer, More Realistic Synthetic Samples from a p-Series Interpolant
Kumar Abhishek,
Colin J. Brown,
Ghassan Hamarneh
Journal of Big Data,
2024
We propose a generalization of mixup with provably and demonstrably desirable properties that
allows convex combinations of more than 2 samples.
[Abstract]
Modern deep learning training procedures rely on model regularization techniques such as
data augmentation methods, which generate training samples that increase the diversity of
data and richness of label information. A popular recent method, mixup, uses convex
combinations of pairs of original samples to generate new samples. However, as we show in our
experiments, mixup can produce undesirable synthetic samples, where the data is sampled
off the manifold and can contain incorrect labels. We propose ζ-mixup, a generalization of
mixup with provably and demonstrably desirable properties that allows convex combinations
of T ≥ 2 samples, leading to more realistic and diverse outputs that incorporate information
from T original samples by using a p-series interpolant. We show that, compared to
mixup, ζ-mixup better preserves the intrinsic dimensionality of the original datasets, which
is a desirable property for training generalizable models. Furthermore, we show that our
implementation of ζ-mixup is faster than mixup, and extensive evaluation on controlled
synthetic and 26 diverse real-world natural and medical image classification datasets shows
that ζ-mixup outperforms mixup, CutMix, and traditional data augmentation techniques.
|
|
Review
Paper
|
A Survey on Deep Learning for Skin Lesion Segmentation
Kumar Abhishek*,
Zahra Mirikharaji*,
Alceu Bissoto,
Catarina Barata,
Sandra Avila,
Eduardo Valle,
M. Emre Celebi,
Ghassan Hamarneh [*: Joint first authors]
Medical Image Analysis,
2023
We review the literature on deep learning-based skin lesion segmentation, evaluating the
current research along several dimensions: input data, model design, and evaluation, and
discuss their limitations and potential research directions.
[Abstract]
Skin cancer is a major public health problem that could benefit from computer-aided
diagnosis to reduce the burden of this common disease. Skin lesion segmentation from
images is an important step toward achieving this goal. However, the presence of
natural and artificial artifacts (e.g., hair and air bubbles), intrinsic factors (e.g., lesion
shape and contrast), and variations in image acquisition conditions make skin lesion
segmentation a challenging task. Recently, various researchers have explored the
applicability of deep learning models to skin lesion segmentation. In this survey, we
cross-examine 177 research papers that deal with deep learning-based segmentation
of skin lesions. We analyze these works along several dimensions, including input data
(datasets, preprocessing, and synthetic data generation), model design (architecture,
modules, and losses), and evaluation aspects (data annotation requirements and
segmentation performance). We discuss these dimensions both from the viewpoint of
select seminal works, and from a systematic viewpoint, examining how those choices
have influenced current trends, and how their limitations should be addressed. To
facilitate comparisons, we summarize all examined works in a comprehensive table as
well as an interactive table available online.
|
|
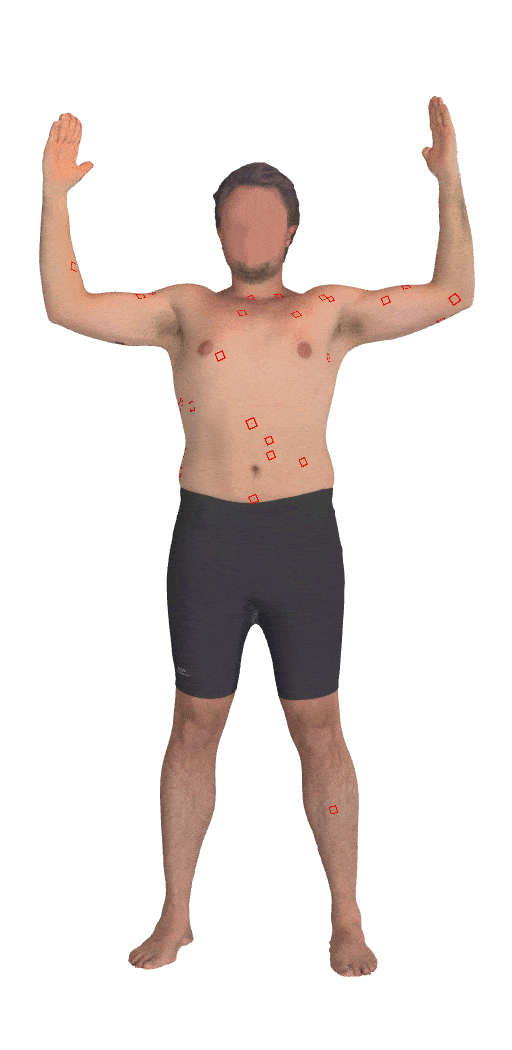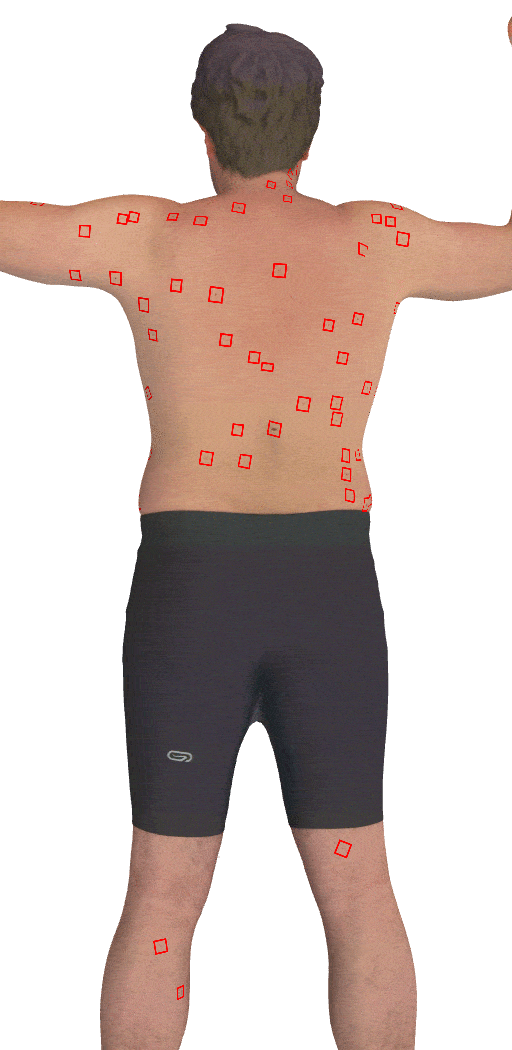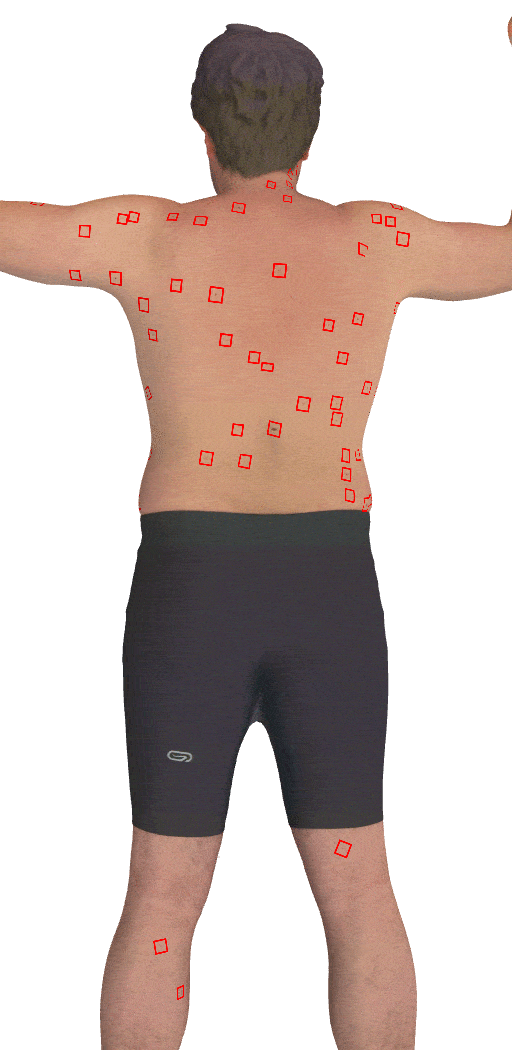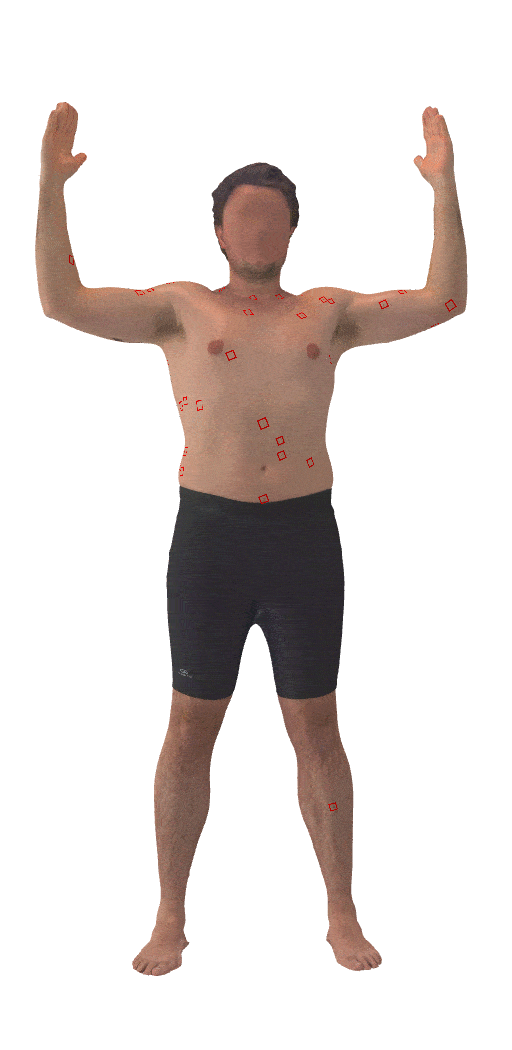
Skin3D: Detection and Longitudinal Tracking of Pigmented Skin Lesions in 3D Total-Body Textured Meshes
Mengliu Zhao*,
Jeremy Kawahara*,
Kumar Abhishek,
Sajjad Shamanian,
Ghassan Hamarneh [*: Joint first authors]
Medical Image Analysis,
2022
We present a deep learning-based approach to detect and track skin lesions on 3D whole-body scans.
[Abstract]
We present an automated approach to detect and longitudinally track skin lesions on 3D total-body
skin surfaces scans. The acquired 3D mesh of the subject is unwrapped to a 2D texture image, where a
trained region convolutional neural network (R-CNN) localizes the lesions within the 2D domain. These
detected skin lesions are mapped back to the 3D surface of the subject and, for subjects imaged
multiple times, the anatomical correspondences among pairs of meshes and the geodesic distances among
lesions are leveraged in our longitudinal lesion tracking algorithm.We evaluated the proposed approach
using three sources of data. Firstly, we augmented the 3D meshes of human subjects from the public
FAUST dataset with a variety of poses, textures, and images of lesions. Secondly, using a handheld
structured light 3D scanner, we imaged a mannequin with multiple synthetic skin lesions at selected
location and with varying shapes, sizes, and colours. Finally, we used 3DBodyTex, a publicly available
dataset composed of 3D scans imaging the colored (textured) skin of 200 human subjects. We manually
annotated locations that appeared to the human eye to contain a pigmented skin lesion as well as
tracked a subset of lesions occurring on the same subject imaged in different poses. Our results, on
test subjects annotated by three human annotators, suggest that the trained R-CNN detects lesions at a
similar performance level as the human annotators. Our lesion tracking algorithm achieves an average
accuracy of 80% when identifying corresponding pairs of lesions across subjects imaged in different
poses. As there currently is no other large-scale publicly available dataset of 3D total-body skin
lesions, we publicly release the 10 mannequin meshes and over 25,000 3DBodyTex manual annotations,
which we hope will further research on total-body skin lesion analysis.
|
|
|
Predicting the Clinical Management of Skin Lesions using Deep Learning
Kumar Abhishek,
Jeremy Kawahara,
Ghassan Hamarneh
Nature Scientific Reports,
2021
We present a deep learning-based approach to predict the clinical management decisions for skin lesions
from images without explicitly predicting the underlying diagnosis.
[Abstract]
Automated machine learning approaches to skin lesion diagnosis from images are approaching
dermatologist-level performance. However, current machine learning approaches that suggest management
decisions rely on predicting the underlying skin condition to infer a management decision without
considering the variability of management decisions that may exist within a single condition. We
present the first work to explore image-based prediction of clinical management decisions directly
without explicitly predicting the diagnosis. In particular, we use clinical and dermoscopic images of
skin lesions along with patient metadata from the Interactive Atlas of Dermoscopy dataset (1,011
cases; 20 disease labels; 3 management decisions) and demonstrate that predicting management labels
directly is more accurate than predicting the diagnosis and then inferring the management decision
(13.73 ± 3.93% and 6.59 ± 2.86% improvement in overall accuracy and AUROC respectively),
statistically significant at p
< 0.001. Directly predicting management decisions also considerably reduces the over-excision rate as
compared to management decisions inferred from diagnosis predictions (24.56% fewer cases wrongly
predicted to be excised). Furthermore, we show that training a model to also simultaneously predict
the seven-point criteria and the diagnosis of skin lesions yields an even higher accuracy
(improvements of 4.68 ± 1.89% and 2.24 ± 2.04% in overall accuracy and AUROC respectively)
of management predictions. Finally, we demonstrate our model's generalizability by evaluating on the
publicly available MClass-D dataset and show that our model agrees with the clinical management
recommendations of 157 dermatologists as much as they agree amongst each other.
|
|
Review
Paper
|
Deep Semantic Segmentation of Natural and Medical Images: A Review
Kumar Abhishek*, Saeid
Asgari
Taghanaki*, Joseph Paul Cohen,
Julien
Cohen-Adad,
Ghassan Hamarneh [*: Joint first authors]
Artificial Intelligence Review, 2021
We present a comprehensive survey of advances in deep learning-based semantic segmentation of natural
and medical images, categorizing the contributions in 6 broad categories, and discuss limitations and
potential research directions.
[Abstract]
The semantic image segmentation task consists of classifying each pixel of an image into an instance,
where each instance corresponds to a class. This task is a part of the concept of scene understanding
or better explaining the global context of an image. In the medical image analysis domain, image
segmentation can be used for image-guided interventions, radiotherapy, or improved radiological
diagnostics. In this review, we categorize the leading deep learning-based medical and non-medical
image segmentation solutions into six main groups of deep architectural, data synthesis-based, loss
function-based, sequenced models, weakly supervised, and multi-task methods and provide a
comprehensive review of the contributions in each of these groups. Further, for each group, we analyze
each variant of these groups and discuss the limitations of the current approaches and present
potential future research directions for semantic image segmentation.
|
|
Review
Paper
|
Artificial Intelligence In Glioma Imaging: Challenges and Advances
Weina Jin,
Mostafa
Fatehi,
Kumar Abhishek,
Mayur Mallya,
Brian Toyota,
Ghassan Hamarneh
Journal of Neural Engineering, 2020
We review the literature to analyze the most important challenges in the clinical adoption of AI-based
methods and present a summary of the recent advances, categorizing them into three broad categories:
dealing with limited data volume and annotations, training of deep learning-based models, and the
clinical deployment of these models.
[Abstract]
Primary brain tumors including gliomas continue to pose significant management challenges to
clinicians. While the presentation, the pathology, and the clinical course of these lesions are
variable, the initial investigations are usually similar. Patients who are suspected to have a brain
tumor will be assessed with computed tomography (CT) and magnetic resonance imaging (MRI). The
imaging findings are used by neurosurgeons to determine the feasibility of surgical resection and
plan such an undertaking. Imaging studies are also an indispensable tool in tracking tumor
progression or its response to treatment. As these imaging studies are non-invasive, relatively
cheap and accessible to patients, there have been many efforts over the past two decades to increase
the amount of clinically-relevant information that can be extracted from brain imaging. Most
recently, artificial intelligence (AI) techniques have been employed to segment and characterize
brain tumors, as well as to detect progression or treatment-response. However, the clinical utility
of such endeavours remains limited due to challenges in data collection and annotation, model
training, and the reliability of AI-generated information.
We provide a review of recent advances in addressing the above challenges. First, to overcome the
challenge of data paucity, different image imputation and synthesis techniques along with annotation
collection efforts are summarized. Next, various training strategies are presented to meet multiple
desiderata, such as model performance, generalization ability, data privacy protection, and learning
with sparse annotations. Finally, standardized performance evaluation and model interpretability
methods have been reviewed. We believe that these technical approaches will facilitate the
development of a fully-functional AI tool in the clinical care of patients with gliomas.
|
|
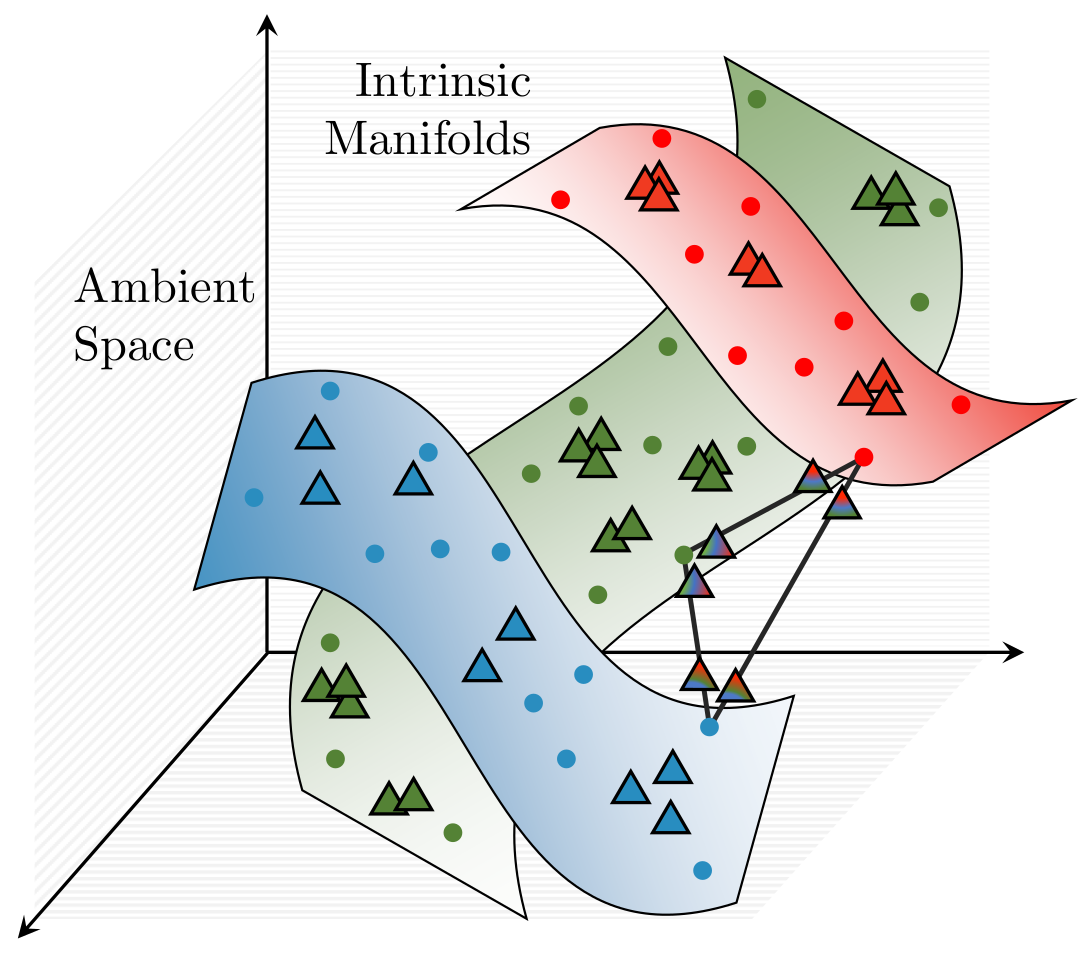
ζ-mixup: Richer, More Realistic Mixing of Multiple Images
Kumar Abhishek,
Colin J. Brown,
Ghassan Hamarneh
Medical Imaging with Deep Learning (MIDL) Short Paper,
2023
We present a multi-sample Riemann zeta-weighted mixing-based image augmentation to
generate richer and more realistic outputs.
[Abstract]
Data augmentation (DA), an effective regularization technique, generates training samples to
enhance the diversity of data and the richness of label information for training modern deep
learning models. mixup, a popular recent DA method, augments training datasets with convex
combinations of original samples pairs, but can generate undesirable samples, with data being
sampled off the manifold and with incorrect labels. In this work, we propose ζ-mixup, a
generalization of mixup with provably and demonstrably desirable properties that allows
for convex combinations of N ≥ 2 samples, thus leading to more realistic and
diverse outputs that incorporate information from N original samples using a
p-series interpolant. We show that, compared to mixup, ζ-mixup better preserves
the intrinsic dimensionality of the original datasets, a desirable property for training
generalizable models, and is at least as fast as mixup. Evaluation on several natural and
medical image datasets shows that ζ-mixup outperforms mixup, CutMix, and traditional
DA methods.
|
|
CIRCLe: Color Invariant Representation Learning for Unbiased Classification of Skin Lesions
Arezou Pakzad,
Kumar Abhishek,
Ghassan Hamarneh
ISIC Skin Image Analysis Workshop, European Conference on Computer Vision (ECCV), 2022
We propose a skin color transformer, a domain invariant representation learning
method, and a new fairness metric for mitigating skin type bias in clinical image
classification.
[Abstract]
While deep learning based approaches have demonstrated expert-level performance in
dermatological diagnosis tasks, they have also been shown to exhibit biases toward
certain demographic attributes, particularly skin types (e.g., light versus dark),
a fairness concern that must be addressed. We propose CIRCLe, a skin color invariant
deep representation learning method for improving fairness in skin lesion classification.
CIRCLe is trained to classify images by utilizing a regularization loss that encourages
images with the same diagnosis but different skin types to have similar latent
representations. Through extensive evaluation and ablation studies, we demonstrate
CIRCLe's superior performance over the state-of-the-art when evaluated on 16k+ images
spanning 6 Fitzpatrick skin types and 114 diseases, using classification accuracy, equal
opportunity difference (for light versus dark groups), and normalized accuracy range, a
new measure we propose to assess fairness on multiple skin type groups.
|
|
|
D-LEMA: Deep Learning Ensembles from Multiple Annotations -- Application to Skin Lesion
Segmentation
Zahra Mirikharaji,
Kumar Abhishek, Saeed Izadi, Ghassan Hamarneh
ISIC Skin Image Analysis Workshop, IEEE International Conference on Computer Vision and Pattern
Recognition (CVPR),
2021 (Best Paper Award)
We propose an ensemble of Bayesian FCNs to perform segmentation from multiple (contradictory)
annotations and fuse predictions from multiple base models to improve confidence calibration.
[Abstract]
Medical image segmentation annotations suffer from inter/intra-observer variations even among experts
due to intrinsic differences in human annotators and ambiguous boundaries. Leveraging a collection of
annotators' opinions for an image is an interesting way of estimating a gold standard. Although
training deep models in a supervised setting with a single annotation per image has been extensively
studied, generalizing their training to work with data sets containing multiple annotations per image
remains a fairly unexplored problem. In this paper, we propose an approach to handle annotators'
disagreements when training a deep model. To this end, we propose an ensemble of Bayesian fully
convolutional networks (FCNs) for the segmentation task by considering two major factors in the
aggregation of multiple ground truth annotations: (1) handling contradictory annotations in the
training data originating from inter-annotator disagreements and (2) improving confidence calibration
through the fusion of base models predictions. We demonstrate the superior performance of our approach
on the ISIC Archive and explore the generalization performance of our proposed method by cross-data
set evaluation on the PH2 and DermoFit data sets.
|
|
|
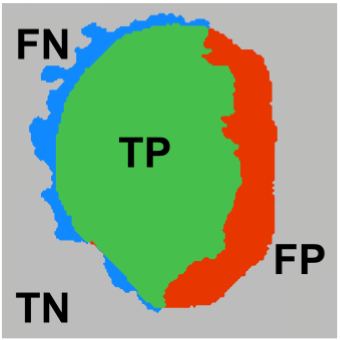
Matthews Correlation Coefficient Loss for Deep Convolutional Networks: Application to Skin
Lesion Segmentation
Kumar Abhishek, Ghassan Hamarneh
International Symposium on Biomedical Imaging (ISBI),
2021
We propose a new overlap-based loss function for binary segmentation that takes into account the true
negative pixels and achieves a better sensitivity-specificity trade-off than the popular Dice loss.
[Abstract]
The segmentation of skin lesions is a crucial task in clinical decision support systems for the
computer aided diagnosis of skin lesions. Although deep learning-based approaches have improved
segmentation performance, these models are often susceptible to class imbalance in the data,
particularly, the fraction of the image occupied by the background healthy skin. Despite variations of
the popular Dice loss function being proposed to tackle the class imbalance problem, the Dice loss
formulation does not penalize misclassifications of the background pixels. We propose a novel
metric-based loss function using the Matthews correlation coefficient, a metric that has been shown to
be efficient in scenarios with skewed class distributions, and use it to optimize deep segmentation
models. Evaluations on three skin lesion image datasets: the ISBI ISIC 2017 Skin Lesion Segmentation
Challenge dataset, the DermoFit Image Library, and the PH2 dataset, show that models trained using the
proposed loss function outperform those trained using Dice loss by 11.25%, 4.87%, and 0.76%
respectively in the mean Jaccard index. The code is available on GitHub.
|
|
|
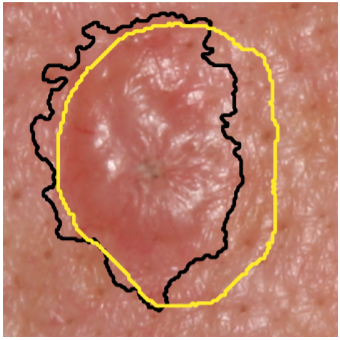
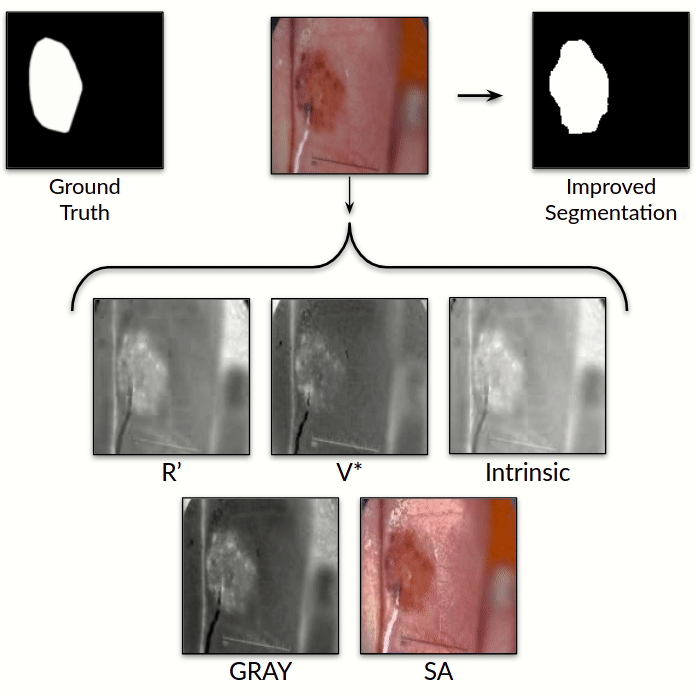
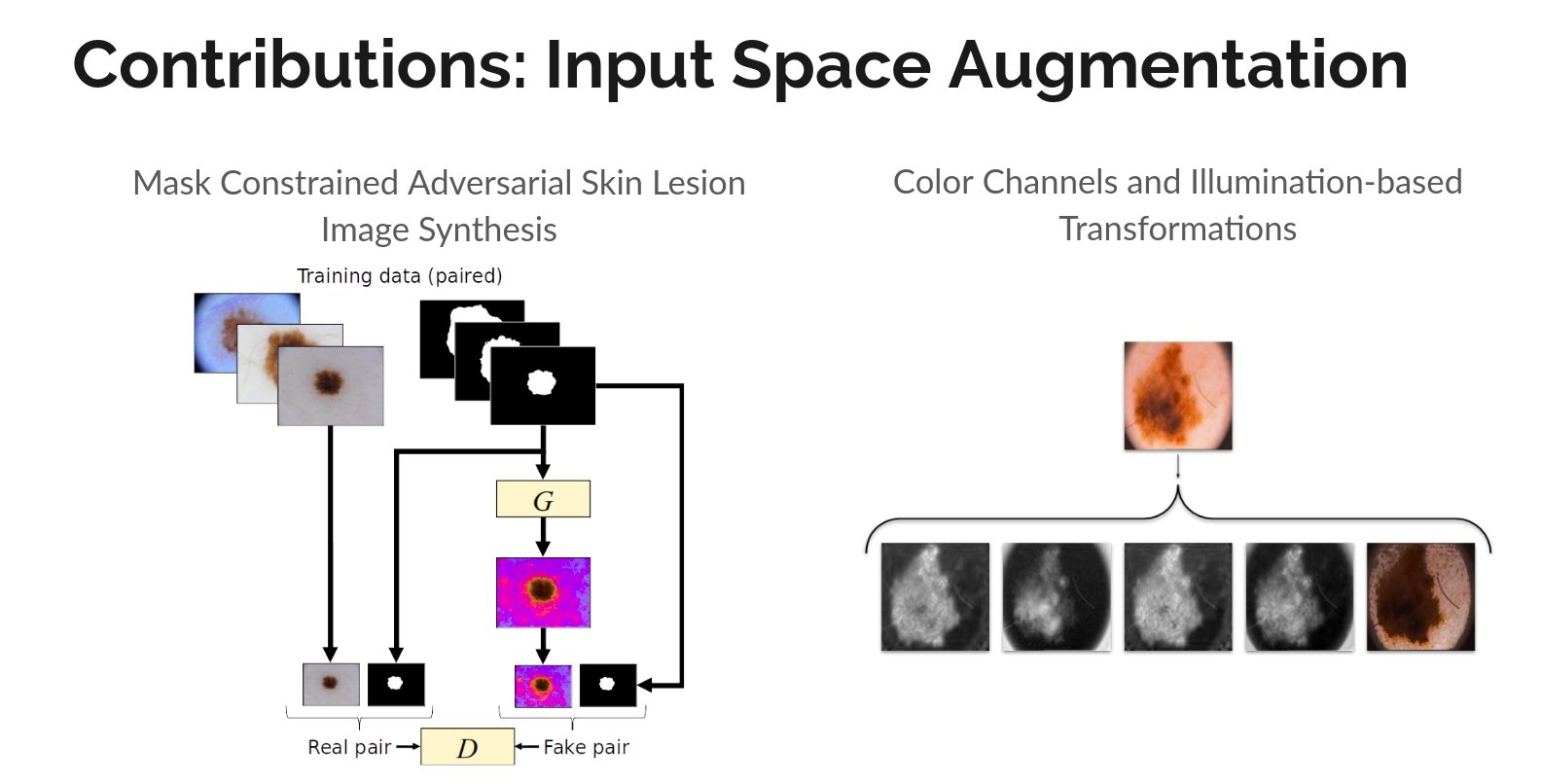
Illumination-based Transformations Improve Skin Lesion Segmentation in Dermoscopic Images
Kumar Abhishek, Ghassan Hamarneh, Mark S. Drew
ISIC Skin Image Analysis Workshop, IEEE International Conference on Computer Vision and Pattern
Recognition (CVPR),
2020
We incorporate information from specific color bands, illumination invariant grayscale images, and
shading-attenuated images obtained from RGB dermoscopic images of skin lesions to improve the lesion
segmentation.
[Abstract]
The semantic segmentation of skin lesions is an important and common initial task in the computer
aided diagnosis of dermoscopic images. Although deep learning-based approaches have considerably
improved the segmentation accuracy, there is still room for improvement by addressing the major
challenges, such as variations in lesion shape, size, color and varying levels of contrast. In this
work, we propose the first deep semantic segmentation framework for dermoscopic images which
incorporates, along with the original RGB images, information extracted using the physics of skin
illumination and imaging. In particular, we incorporate information from specific color bands,
illumination invariant grayscale images, and shading-attenuated images. We evaluate our method on
three datasets: the ISBI ISIC 2017 Skin Lesion Segmentation Challenge dataset, the DermoFit Image
Library, and the PH2 dataset and observe improvements of 12.02%, 4.30%, and 8.86% respectively in
the mean Jaccard index over a baseline model trained only with RGB images.
|
|
|
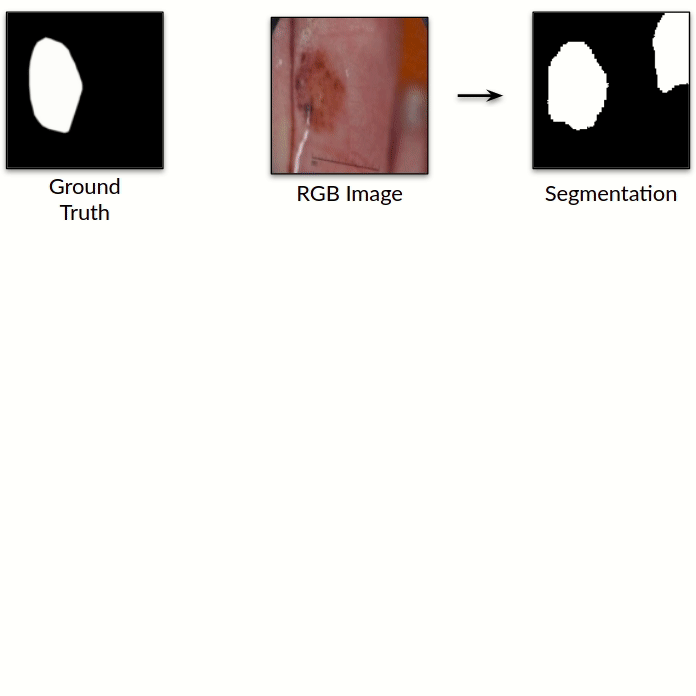
Mask2Lesion: Mask-Constrained Adversarial Skin Lesion Image Synthesis
Kumar Abhishek, Ghassan Hamarneh
Workshop on Simulation and Synthesis in Medical Imaging (SASHIMI), International Conference on
Medical Image Computing and Computer Assisted Intervention (MICCAI),
2019
We propose a GAN-based synthesis approach for generating realistic skin lesion images from lesion
masks, making it an appropriate augmentation strategy for skin lesion segmentation datasets.
[Abstract]
[bibtex]
Skin lesion segmentation is a vital task in skin cancer diagnosis and further treatment. Although
deep learning based approaches have significantly improved the segmentation accuracy, these
algorithms are still reliant on having a large enough dataset in order to achieve adequate results.
Inspired by the immense success of generative adversarial networks (GANs), we propose a GAN-based
augmentation of the original dataset in order to improve the segmentation performance. In
particular, we use the segmentation masks available in the training dataset to train the Mask2Lesion
model, and use the model to generate new lesion images given any arbitrary mask, which are then used
to augment the original training dataset. We test Mask2Lesion augmentation on the ISBI ISIC 2017
Skin Lesion Segmentation Challenge dataset and achieve an improvement of 5.17% in the mean Dice
score as compared to a model trained with only classical data augmentation techniques.
|
|
|
Improved Inference via Deep Input Transfer
Saeid Asgari Taghanaki, Kumar
Abhishek, Ghassan Hamarneh
International Conference on Medical Image Computing and Computer Assisted Intervention
(MICCAI), 2019 (Early Accept)
We propose an input image transformation technique that relies on the gradients of a trained
segmentation network to transform the images for improved segmentation performance.
[Abstract]
[bibtex]
Although numerous improvements have been made in the field of image segmentation using
convolutional neural networks, the majority of these improvements rely on training with larger
datasets, model architecture modifications, novel loss functions, and better optimizers. In this
paper, we propose a new segmentation performance boosting paradigm that relies on optimally
modifying the network's input instead of the network itself. In particular, we leverage the
gradients of a trained segmentation network with respect to the input to transfer it to a space
where the segmentation accuracy improves. We test the proposed method on three publicly available
medical image segmentation datasets: the ISIC 2017 Skin Lesion Segmentation dataset, the Shenzhen
Chest X-Ray dataset, and the CVC-ColonDB dataset, for which our method achieves improvements of
5.8%, 0.5%, and 4.8% in the average Dice scores, respectively.
|
|
|
CloudMaskGAN: A Content-Aware Unpaired Image-to-Image Translation Algorithm for Remote
Sensing Imagery
Sorour Mohajerani, Reza Asad, Kumar Abhishek, Neha Sharma, Alysha van
Duynhoven, Parvaneh Saeedi
IEEE International Conference on Image Processing (ICIP), 2019
We propose an unpaired image-to-image translation algorithm for generating synthetic remote sensing
images with different land cover types while preserving the locations and the intensity values of the
cloud pixels.
[Abstract]
[bibtex]
Cloud segmentation is a vital task in applications that utilize satellite imagery. A common
obstacle in using deep learning-based methods for this task is the lack of a large number of images
with their annotated ground truths. This work presents a content-aware unpaired image-to-image
translation algorithm. It generates synthetic images with different land cover types from original
images, while it preserves the locations and the intensity values of the cloud pixels. Therefore, no
manual annotation of ground truth in these images is required. The visual and numerical evaluations
of the generated images by the proposed method prove that their quality is better than that of
competitive algorithms.
|
|
|
A Kernelized Manifold Mapping to Diminish the Effect of Adversarial Perturbations
Saeid Asgari Taghanaki, Kumar
Abhishek, Shekoofeh Azizi, Ghassan Hamarneh
IEEE International Conference on Computer Vision and Pattern Recognition (CVPR), 2019
We propose a non-linear radial basis convolutional feature mapping based adversarial defense that is
resilient to gradient and non-gradient based attacks while also not affecting the performance of clean
data.
[Abstract]
[bibtex]
The linear and non-flexible nature of deep convolutional models makes them vulnerable to carefully
crafted adversarial perturbations. To tackle this problem, we propose a non-linear radial basis
convolutional feature mapping by learning a Mahalanobis-like distance function. Our method then maps
the convolutional features onto a linearly well-separated manifold, which prevents small adversarial
perturbations from forcing a sample to cross the decision boundary. We test the proposed method on
three publicly available image classification and segmentation datasets namely, MNIST, ISBI ISIC
2017 skin lesion segmentation, and NIH Chest X-Ray-14. We evaluate the robustness of our method to
different gradient (targeted and untargeted) and non-gradient based attacks and compare it to
several non-gradient masking defense strategies. Our results demonstrate that the proposed method
can increase the resilience of deep convolutional neural networks to adversarial perturbations
without accuracy drop on clean data.
|
|
Pre-prints and Older Publications
|
|
Review
Paper
|
Attribution-based XAI Methods in Computer Vision: A Review
Kumar Abhishek*,
Deeksha Kamath* [*: Joint first
authors]
arXiv pre-print, arXiv:2211.14736,
2020
We review the current literature in attribution-based XAI methods for computer vision,
particularly gradient-based, perturbation-based, and contrastive methods for XAI, and
discuss the key challenges in developing and evaluating robust XAI methods.
[Abstract]
The advancements in deep learning-based methods for visual perception tasks have seen
astounding growth in the last decade, with widespread adoption in a plethora of
application areas from autonomous driving to clinical decision support systems. Despite
their impressive performance, these deep learning-based models remain fairly opaque in
their decision-making process, making their deployment in human-critical tasks a risky
endeavor. This in turn makes understanding the decisions made by these models crucial
for their reliable deployment. Explainable AI (XAI) methods attempt to address this by
offering explanations for such black-box deep learning methods. In this paper, we
provide a comprehensive survey of attribution-based XAI methods in computer vision and
review the existing literature for gradient-based, perturbation-based, and contrastive
methods for XAI, and provide insights on the key challenges in developing and evaluating
robust XAI methods.
|
|
|
Signed Input Regularization
Kumar Abhishek*, Saeid
Asgari
Taghanaki*, Ghassan Hamarneh [*: Joint first
authors]
arXiv pre-print, arXiv:1911.07086,
2019
We propose a new regularization technique which learns to estimate the contribution of the input
variables in the final prediction output and can be used as a data augmentation strategy.
[Abstract]
[bibtex]
Over-parameterized deep models usually over-fit to a given training distribution, which makes them
sensitive to small changes and out-of-distribution samples at inference time, leading to low
generalization performance. To this end, several model-based and randomized data-dependent
regularization methods are applied, such as data augmentation, which prevent a model from memorizing
the training distribution. Instead of the random transformation of the input images, we propose
SIGN, a new regularization method, which modifies the input variables using a linear transformation
by estimating each variable's contribution to the final prediction. Our proposed technique maps the
input data to a new manifold where the less important variables are de-emphasized. To test the
effectiveness of the proposed idea and compare it with other competing methods, we design several
test scenarios, such as classification performance, uncertainty, out-of-distribution, and robustness
analyses. We compare the methods using three different datasets and four models. We find that SIGN
encourages more compact class representations, which results in the model's robustness to random
corruptions and out-of-distribution samples while also simultaneously achieving superior performance
on normal data compared to other competing methods. Our experiments also demonstrate the successful
transferability of the SIGN samples from one model to another.
|
|
|
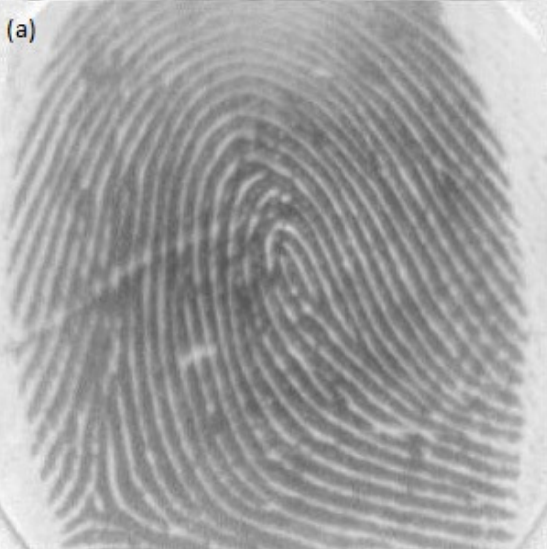
A Minutiae Count Based Method for Fake Fingerprint Detection
Kumar Abhishek, Ashok Yogi
Procedia Computer Science, Volume 58, 2015
[Abstract] [bibtex]
Fingerprint based biometric systems are ubiquitous because they are relatively cheaper to install
and maintain, while serving as a fairly accurate biometric trait. However, it has been shown in the
past that spoofing attacks on many fingerprint scanners are possible using artificial fingerprints
generated using, but not limited to gelatin, Play-Doh and Silicone molds. In this paper, we propose
a novel method based on the minutiae count for detecting the fake fingerprints generated using these
methods. The proposed algorithm has been tested on the standard FVC (Fingerprint Verification
Competition) 2000-2006 dataset and the accuracy was reported to be well above 85%. We also present a
literature survey of the previous algorithms for fake fingerprint detection.
|
|
Non-Invasive Measurement of Heart Rate and Hemoglobin Concentration through Fingertip
Kumar Abhishek, Amodh Kant
Saxena, Ramesh Kumar Sonkar
IEEE International Conference on Signal Processing, Informatics, Communication and Energy Systems
(SPICES), 2015 (Oral Presentation)
[Abstract] [Presentation] [bibtex]
This paper proposes a low cost method for the estimation of blood parameters using
photo-plethysmography (PPG) technique. Data is obtained non-invasively from the fingertip, which is
processed to estimate the heart rate and the hemoglobin concentration. The signal received from
fingertip is first processed via analog filters and the output is sent to a computer through a
microcontroller interface to be processed using MATLAB in order to estimate the parameters. The
proposed method can be used for health monitoring purposes in rural areas. Furthermore, this
solution could be ported to work on a standalone device, thus eliminating the need for a computer.
|
|
|
An Enhanced Algorithm for the Quantification of Human Chorionic Gonadotropin (hCG) Level
in Commercially Available Home Pregnancy Test Kits
Kumar Abhishek, Mrinal Haloi, Sumohana S. Channappayya, Siva Rama Krishna Vanjari, Dhananjaya Dendukuri, Swathy Sridharan, Tripurari
Choudhary, Paridhi
Bhandari
IEEE Twentieth National Conference on Communications (NCC), 2014 (Oral Presentation)
[Abstract] [Presentation] [bibtex]
Home pregnancy kits typically provide a qualitative (yes/no) result based on the concentration of
human chorionic gonadotropin (hCG) present in urine samples. We present an algorithm that converts
this purely qualitative test into a semiquantitative one by processing digital images of the test
kit's output. The algorithm identifies the test and control lines in the image and classifies an
input into one of four different hCG concentration levels based on the color of the test line. The
proposed algorithm provides significant improvement over a prior method and reduces the maximum
false positive rate to less than 5%. This improvement is achieved by a careful choice of the color
space so as to maximize the inter-concentration separability. Also, the proposed method increases
the utility of the test kits by providing useful diagnostic information. Furthermore, the algorithm
could be ported to a mobile platform to make it particularly helpful in remote rural health
monitoring.
|
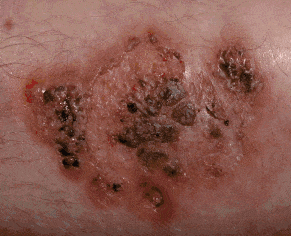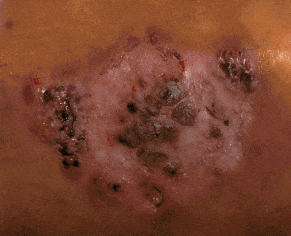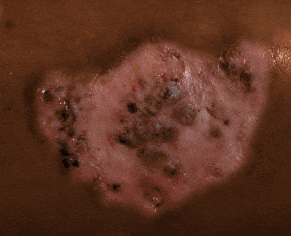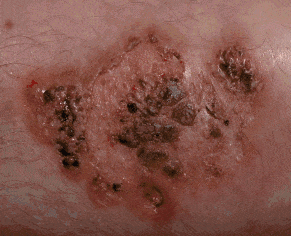
|
Input Space Augmentation for Skin Lesion Segmentation in Dermoscopic Images
Master's Thesis
[Abstract]
Cancer is the second leading cause of death globally, and of all the cancers, skin cancer
is the most prevalent. Early diagnosis of skin cancer is a crucial step for maximizing patient
survival rates and treatment outcomes. Skin conditions are often diagnosed by dermatologists
based on the visual properties of the affected regions, motivating the utility of automated
algorithms to assist dermatologists and offer viable, low-cost, and quick results to assist
dermatological diagnoses. Over the last decade, machine learning, and more recently, deep
learning-based diagnoses of skin lesions have started approaching human performance levels.
This thesis studies approaches to improve the segmentation of skin lesions in dermoscopic
images, which is often the first and the most important task in the diagnosis of dermatological
conditions. In particular, we present two methods to improve deep learning-based segmentation
of skin lesions by augmenting the input space of convolutional neural network models. In the
first contribution, we address the problem of the paucity of annotated data by learning to
synthesize artificial skin lesion images conditioned on input segmentation masks. We then
use these synthetic image mask pairs to augment our original segmentation training datasets.
In our second contribution, we leverage certain color channels and skin imaging- and
illumination-based knowledge in a deep learning framework to augment the input space of the
segmentation models. We evaluate the two contributions on five dermoscopic image datasets:
the ISIC Skin Lesion Segmentation Challenge 2016, 2017, and 2018 datasets, the DermoFit Image
Library, and the PH2 Database, and observe performance improvements across all datasets.
|
|
|
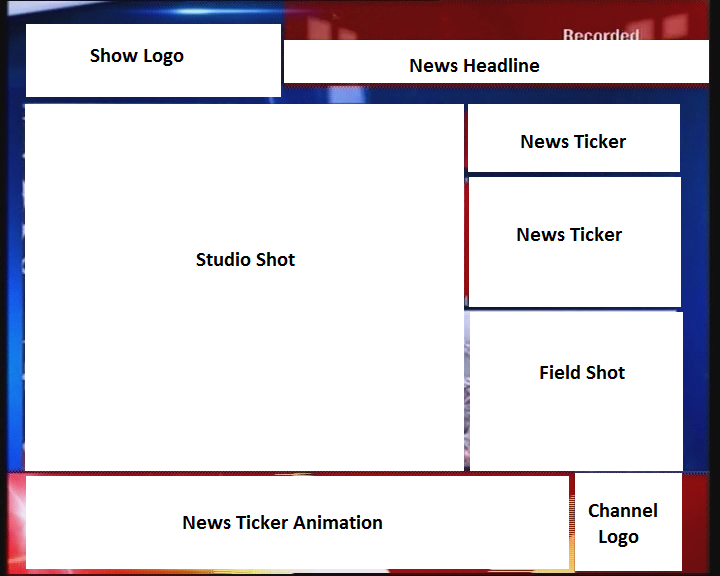
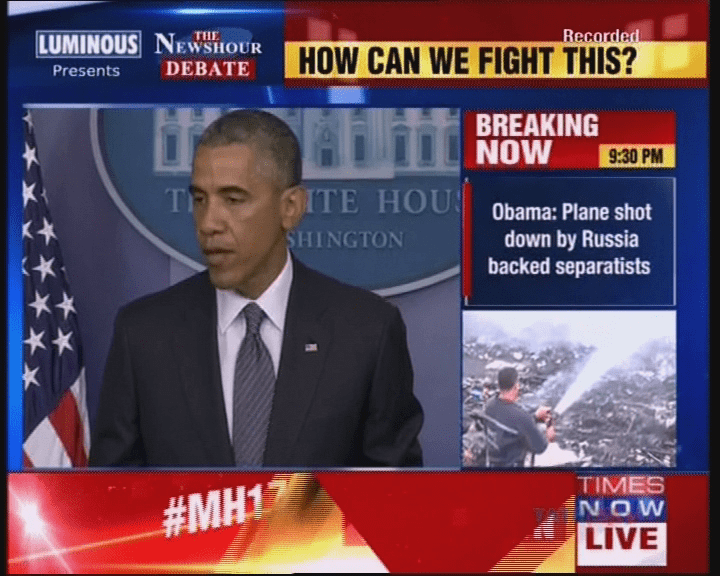
Summarization and Visualization of Large Volumes of Broadcast Video Data
Undergraduate Thesis
[Abstract] [bibtex]
Over the past few years, there has been an astounding growth in the number of news channels as well
as the amount of broadcast news video data. As a result, it is imperative that automated methods
need to be developed in order to effectively summarize and store this voluminous data. Format
detection of news videos plays an important role in news video analysis. Our problem involves
building a robust and versatile news format detector, which identifies the different band elements
in a news frame. Probabilistic progressive Hough transform has been used for the detection of band
edges. The detected bands are classified as natural images, computer generated graphics (non-text)
and text bands. A contrast based text detector has been used to identify the text regions from news
frames. Two classifers have been trained and evaluated for the labeling of the detected bands as
natural or artificial - Support Vector Machine (SVM) Classifer with RBF kernel, and Extreme Learning
Machine (ELM) classifier. The classifiers have been trained on a dataset of 6000 images (3000 images
of each class). The ELM classifier reports a balanced accuracy of 77.38%, while the SVM classifier
outperforms it with a balanced accuracy of 96.5% using 10-fold cross-validation. The detected bands
which have been fragmented due to the presence of gradients in the image have been merged using a
three-tier hierarchical reasoning model. The bands were detected with a Jaccard Index of 0.8138,
when compared to manually marked ground truth data. We have also presented an extensive literature
review of previous work done towards news videos format detection, element band classification, and
associative reasoning.
|

|
Teaching Assistant
CMPT 340: Biomedical Computing
- Spring 2021
- Summer 2021
- Fall 2023
- Spring 2024
|
|
Reviewer
|
Journals
- Medical Image Analysis (MedIA)
- Computer Methods and Programs in Biomedicine (CMPB)
- Computers in Biology and Medicine (CIBM)
- Nature Scientific Reports (Nat Sci Rep)
- Journal of Nuclear Medicine (JNM)
- npj Imaging
Conferences and Workshops
- Medical Image Computing and Computer Assisted Intervention (MICCAI)
- International Skin Imaging Collaboration (ISIC) Skin Image Analysis Workshop
- Information Processing in Medical Imaging (IPMI)
- Medical Imaging Meets NeurIPS (MedNeurIPS)
|
There are two kinds of scientific progress: the methodical experimentation and categorization
which gradually extend the boundaries of knowledge, and the revolutionary leap of genius which
redefines and transcends those boundaries. Acknowledging our debt to the former, we yearn
nonetheless for the latter. - Prokhor Zakharov, Sid Meier's Alpha Centauri
|