Chapter Contents
Previous
Next
|
Chapter Contents |
Previous |
Next |
| The CALIS Procedure |
| Data Set Options | Short Description |
| DATA= | input data set |
| INEST= | input initial values, constraints |
| INRAM= | input model |
| INWGT= | input weight matrix |
| OUTEST= | covariance matrix of estimates |
| OUTJAC | Jacobian into OUTEST= data set |
| OUTRAM= | output model |
| OUTSTAT= | output statistic |
| OUTWGT= | output weight matrix |
| Data Processing | Short Description |
| AUGMENT | analyzes augmented moment matrix |
| COVARIANCE | analyzes covariance matrix |
| EDF= | defines nobs by number error df |
| NOBS= | defines number of observations nobs |
| NOINT | analyzes uncorrected moments |
| RDF= | defines nobs by number regression df |
| RIDGE | specifies ridge factor for moment matrix |
| UCORR | analyzes uncorrected CORR matrix |
| UCOV | analyzes uncorrected COV matrix |
| VARDEF= | specifies variance divisor |
| Estimation Methods | Short Description |
| METHOD= | estimation method |
| ASYCOV= | formula of asymptotic covariances |
| DFREDUCE= | reduces degrees of freedom |
| G4= | algorithm for STDERR |
| NODIAG | excludes diagonal elements from fit |
| WPENALTY= | penalty weight to fit correlations |
| WRIDGE= | ridge factor for weight matrix |
| Optimization Techniques | Short Description |
| TECHNIQUE= | minimization method |
| UPDATE= | update technique |
| LINESEARCH= | line-search method |
| FCONV= | function convergence criterion |
| GCONV= | gradient convergence criterion |
| INSTEP= | initial step length (RADIUS=, SALPHA=) |
| LSPRECISION= | line-search precision (SPRECISION=) |
| MAXFUNC= | max number function calls |
| MAXITER= | max number iterations |
| Displayed Output Options | Short Description |
| KURTOSIS | compute and display kurtosis |
| MODIFICATION | modification indices |
| NOMOD | no modification indices |
| NOPRINT | suppresses the displayed output |
| PALL | all displayed output (ALL) |
| PCORR | analyzed and estimated moment matrix |
| PCOVES | covariance matrix of estimates |
| PDETERM | determination coefficients |
| PESTIM | parameter estimates |
| PINITIAL | pattern and initial values |
| PJACPAT | displays structure of variable and constant |
| elements of the Jacobian matrix | |
| PLATCOV | latent variable covariances, scores |
| PREDET | displays predetermined moment matrix |
| PRIMAT | displays output in matrix form |
| adds default displayed output | |
| PRIVEC | displays output in vector form |
| PSHORT | reduces default output (SHORT) |
| PSUMMARY | displays only fit summary (SUMMARY) |
| PWEIGHT | weight matrix |
| RESIDUAL= | residual matrix and distribution |
| SIMPLE | univariate statistics |
| STDERR | standard errors |
| NOSTDERR | computes no standard errors |
| TOTEFF | displays total and indirect effects |
| Miscellaneous Options | Short Description |
| ALPHAECV= | probability Browne & Cudeck ECV |
| ALPHARMS= | probability Steiger & Lind RMSEA |
| BIASKUR | biased skewness and kurtosis |
| DEMPHAS= | emphasizes diagonal entries |
| FDCODE | uses numeric derivatives for code |
| HESSALG= | algorithm for Hessian |
| NOADJDF | no adjustment of df for active constraints |
| RANDOM= | randomly generated initial values |
| SINGULAR= | singularity criterion |
| ASINGULAR= | absolute singularity information matrix |
| COVSING= | singularity tolerance of information matrix |
| MSINGULAR= | relative M singularity of information matrix |
| VSINGULAR= | relative V singularity of information matrix |
| SLMW= | probability limit for Wald test |
| START= | constant initial values |
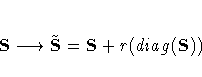
| Value | Description | Divisor |
| DF | degrees of freedom | N - k - i |
| N | number of observations | N |
| WDF | sum of weights DF | |
| WEIGHT | WGT | sum of weights |
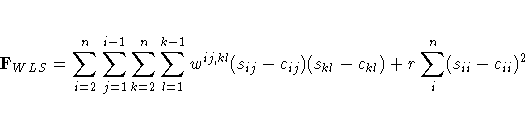
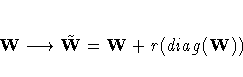
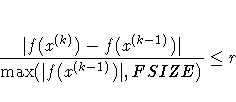
![{ [g(x^{(k)})]^' [G^{(k)}]^{-1} g(x^{(k)}) \over
\max(| f(x^{(k)})|,FSIZE) } \leq r](images/caleq66.gif)
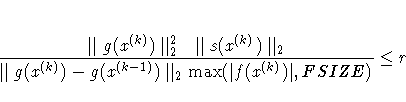
| TECH= | UPDATE= | LSP default |
| QUANEW | DBFGS, BFGS | r = 0.4 |
| QUANEW | DDFP, DFP | r = 0.06 |
| CONGRA | all | r = 0.1 |
| NEWRAP | no update | r = 0.9 |
| TECH= | MAXFUNC default |
| LEVMAR, NEWRAP, NRRIDG, TRUREG | i=125 |
| DBLDOG, QUANEW | i=500 |
| CONGRA | i=1000 |
| TECH= | MAXITER default |
| LEVMAR, NEWRAP, NRRIDG, TRUREG | i=50 |
| DBLDOG, QUANEW | i=200 |
| CONGRA | i=400 |
maxiter= . 0means that you do not want to exceed the default number of iterations during the main optimization process and that you want to suppress the feasible point algorithm for nonlinear constraints.
| Output Options | PALL | default | PSHORT | PSUMMARY | |
| fit indices | * | * | * | * | * |
| linear dependencies | * | * | * | * | * |
| PREDET | * | (*) | (*) | (*) | |
| model matrices | * | * | * | * | |
| PESTIM | * | * | * | * | |
| iteration history | * | * | * | * | |
| PINITIAL | * | * | * | ||
| SIMPLE | * | * | * | ||
| STDERR | * | * | * | ||
| RESIDUAL | * | * | |||
| KURTOSIS | * | * | |||
| PLATCOV | * | * | |||
| TOTEFF | * | * | |||
| PCORR | * | ||||
| MODIFICATION | * | ||||
| PWEIGHT | * | ||||
| PCOVES | |||||
| PDETERM | |||||
| PJACPAT | |||||
| PRIMAT | |||||
| PRIVEC |
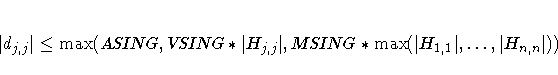
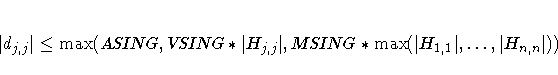
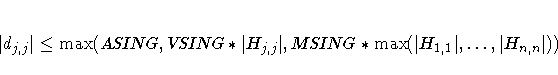
|
Chapter Contents |
Previous |
Next |
Top |
Copyright © 1999 by SAS Institute Inc., Cary, NC, USA. All rights reserved.