| Model Specification Options |
| LINK= | specifies link function |
| NOINT | suppresses intercept |
| NOFIT | suppresses model fitting |
| OFFSET= | specifies offset variable |
| SELECTION= | specifies variable selection method |
| Variable Selection Options |
|
BEST= | controls the number of models displayed for SCORE selection |
| DETAILS | requests detailed results at each step |
| FAST | uses fast elimination method |
| HIERARCHY= | specifies whether and how hierarchy is maintained and whether
a single effect or multiple effects are allowed to enter or leave
the model per step |
| INCLUDE= | specifies number of variables included in every model |
| MAXSTEP= | specifies maximum number of steps for STEPWISE selection |
| SEQUENTIAL | adds or deletes variables in sequential order |
| SLENTRY= | specifies significance level for entering variables |
| SLSTAY= | specifies significance level for removing variables |
| START= | specifies the number of variables in first model |
| STOP= | specifies the number of variables in final model |
| STOPRES | adds or deletes variables by residual chi-square criterion |
| Model-Fitting Specification Options |
| ABSFCONV= | specifies the absolute function convergence criterion |
| FCONV= | specifies the relative function convergence criterion |
| GCONV= | specifies the relative gradient convergence criterion |
| XCONV= | specifies the relative parameter convergence criterion |
| MAXITER= | specifies maximum number of iterations |
| NOCHECK | suppresses checking for infinite parameters |
| RIDGING= | specifies the technique used to improve the log-likelihood function
when its value is worse than that of the previous step |
| SINGULAR= | specifies tolerance for testing singularity |
| TECHNIQUE= | specifies iterative algorithm for maximization |
| Options for Confidence Intervals |
| ALPHA= | specifies 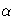 for the for the  confidence intervals confidence intervals |
| CLPARM= | computes confidence intervals for parameters |
| CLODDS= | computes confidence intervals for odds ratios |
| PLCONV= | specifies profile likelihood convergence criterion |
| Options for Classifying Observations |
| CTABLE | displays classification table |
| PEVENT= | specifies prior event probabilities |
| PPROB= | specifies probability cutpoints for classification |
| Options for Overdispersion and Goodness-of-Fit Tests |
| AGGREGATE= | determines subpopulations for Pearson chi-square and deviance |
| SCALE= | species method to correct overdispersion |
| LACKFIT | requests Hosmer and Lemeshow goodness-of-fit test |
| Options for ROC Curves |
| OUTROC= | names the output data set |
| ROCEPS= | specifies probability grouping criterion |
| Options for Regression Diagnostics |
| INFLUENCE | displays influence statistics |
| IPLOTS | requests index plots |
| Options for Display of Details |
| CORRB | displays correlation matrix |
| COVB | displays covariance matrix |
| EXPB | displays the exponentiated values of estimates |
| ITPRINT | displays iteration history |
| NODUMMYPRINT | suppresses the "Class Level Information" table |
| PARMLABEL | displays the parameter labels |
| RSQUARE | displays generalized R2 |
| STB | displays the standardized estimates |





