|
1)Eye-gaze
Driven Interactive Image Segmentation
We
developed a hands-free interactive image segmentation using an
eye-gaze tracking system.
Abstract:
This paper explores a novel approach to interactive
user-guided image segmentation, using eyegaze information as an
input. The method includes three steps: 1) eyegaze tracking for
providing user input, such as setting object and background seed
pixel selection; 2) an optimization method for image labeling
that is constrained or affected by user input; and 3) linking
the two previous steps via a graphical user interface for
displaying the images and other controls to the user and for
providing real-time visual feedback of eyegaze and seed
locations, thus enabling the interactive segmentation procedure.
We developed a new graphical user interface supported by an
eyegaze tracking monitor to capture the user's eyegaze movement
and fixations (as opposed to traditional mouse moving and
clicking). The user simply looks at different parts of the
screen to select which image to segment, to perform foreground
and background seed placement and to set optional segmentation
parameters.
There is an eyegaze-controlled "zoom" feature for difficult
images containing objects with narrow parts, holes or weak
boundaries. The image is then segmented using the random walker
image segmentation method. We performed a pilot study with 7
subjects who segmented synthetic, natural and real medical
images. Our results show that getting used the new interface
takes about only 5 minutes. Compared with traditional
mouse-based control, the new eyegaze approach provided a 18.6%
speed improvement for more than 90% of images with high
object-background contrast. However, for low contrast and more
difficult images it took longer to place seeds using the eyegaze-based
"zoom" to relax the required eyegaze accuracy of seed placement.
Random Walker Segmentation
:
-
The user
species seed points and the seeds are labeled as either
object or background;
-
The image
is modeled as a graph where image pixels are represented
by graph nodes;
-
Graph
edges connect neighboring pixels;
-
The
weight of an edge is set as a function of the intensity
difference between a pixel and its neighbor. This
function or mapping is controlled by a parameter beta;
-
The
probability that a random walker starting from a
particular pixel reaching any of the labeled pixels
(seeds) is computed for every pixel by solving a linear
system of equations;
-
The
maximum probability label is assigned to each pixel,
which constitutes the image segmentation.
 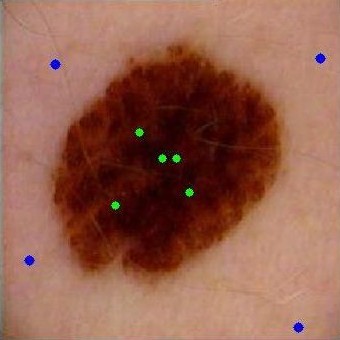  
From
left to right respectively: Original image,
obj and bckG seeds, probability map, and
segmented lesion
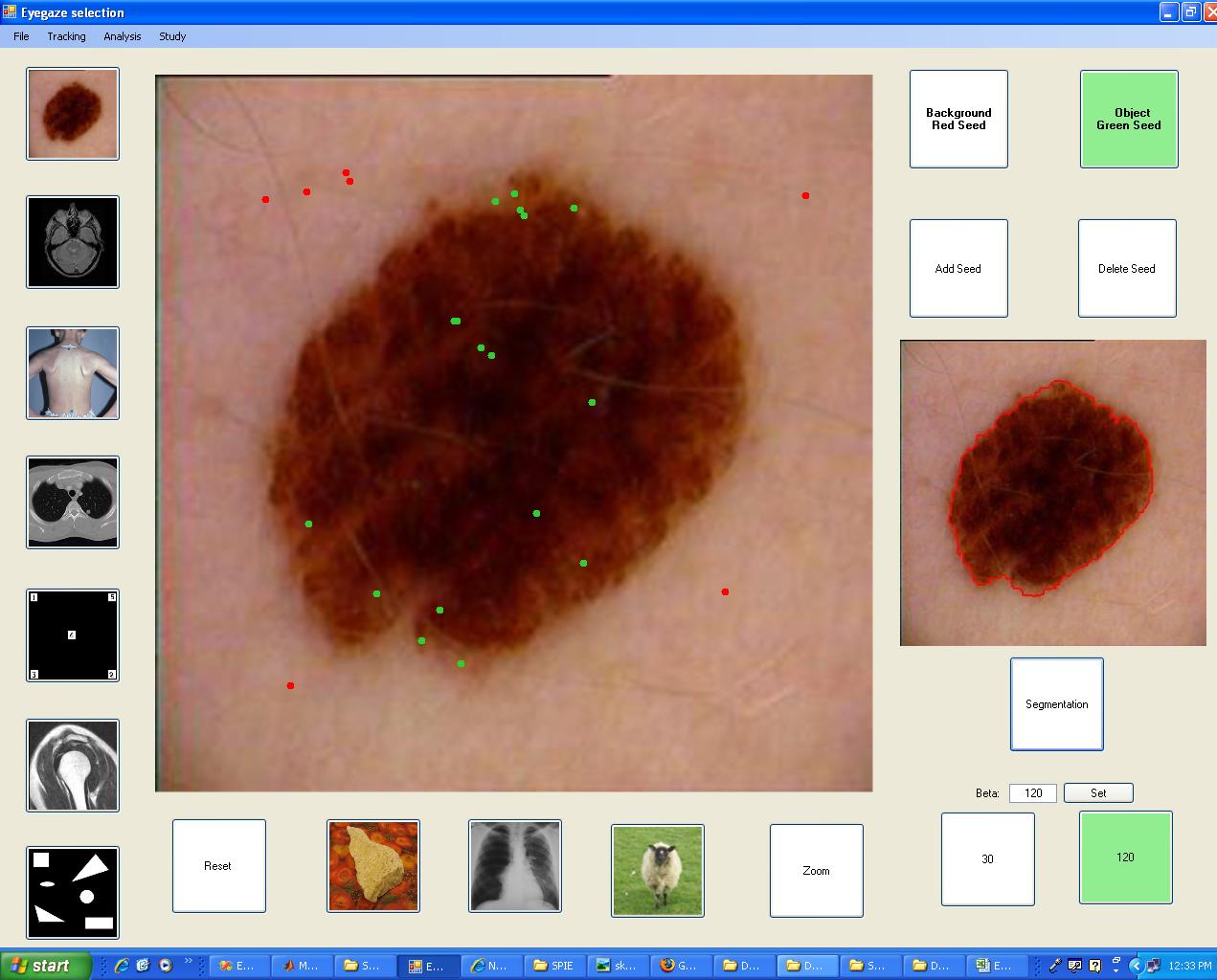
The user-interface used to segment the lesion
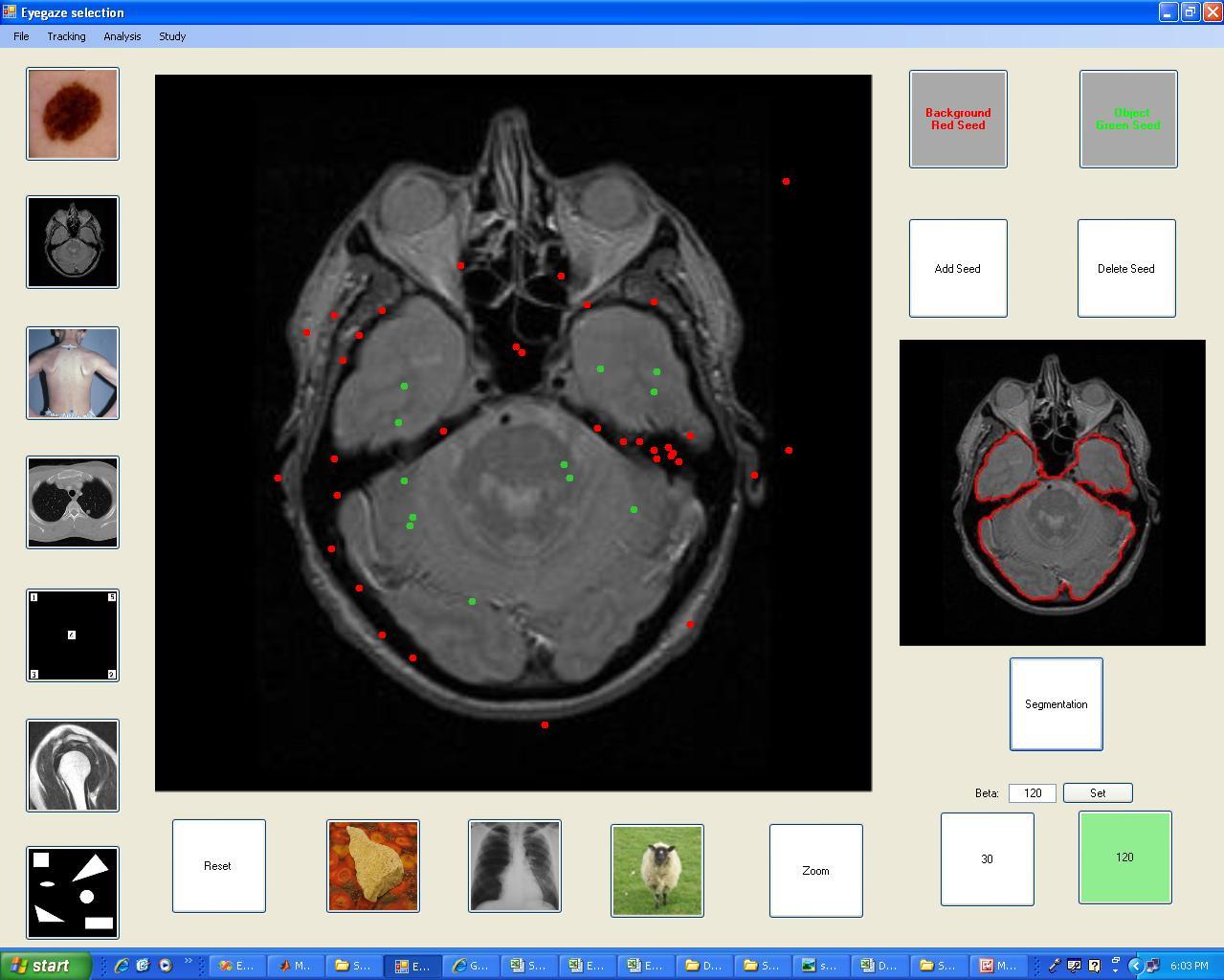 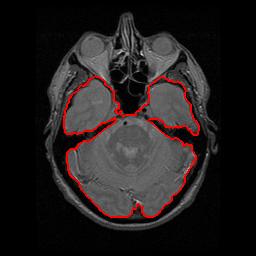
Result of the segmentation on a more
difficult image
Publication (PDF):
Maryam Sadeghi, Geoff Tien, Ghassan Hamarneh, and Stella
Atkins. Hands-free Interactive Image Segmentation Using
Eyegaze. In SPIE Medical Imaging, 2009.
|