Probabilistic models in Automated Skin lesion diagnosis
We are investigating generalized probabilistic models for assigning arbitrary labels to dermoscopic images learnt from an exemplar set . Labels can be either 'image-based' (diagnosis='melanoma', pigment_network='atypical', etc) or 'pixel-based' (segmentation='background', pigment_network='absent', etc.)
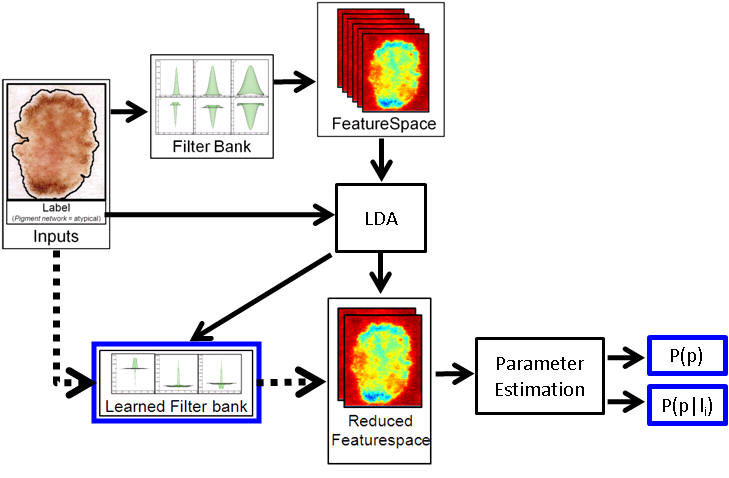
The initial model employs pixel based filter responses as the featurespace. Linear discriminant analysis (LDA) is performed over the featurespace to find the linear combination of features which best separates the labels (and to learn the corresponding filterbank). Distributions of labels over this reduced filterspace are then estimated. Future work will investigate replacing the LDA step with kernel methods.
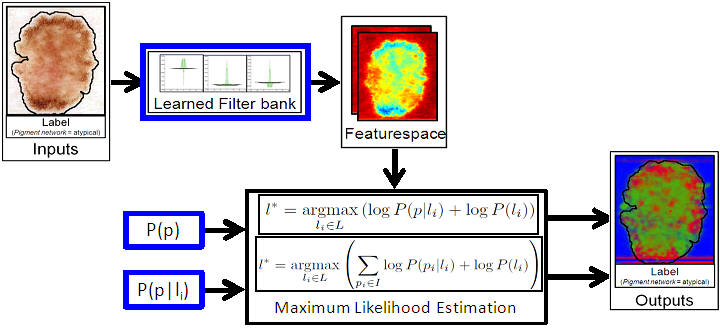
Given an unseen image, the learnt filterbank is used to compute the reduced featureset. Maximum likelihood estimation is then used to assign labels to individual pixels, as well as the entire image. Future work will investigate employing conditional random fields to assign labels.
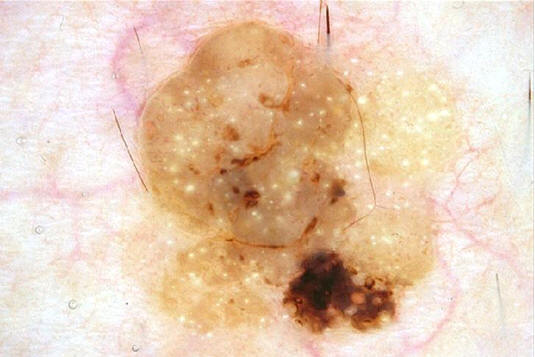
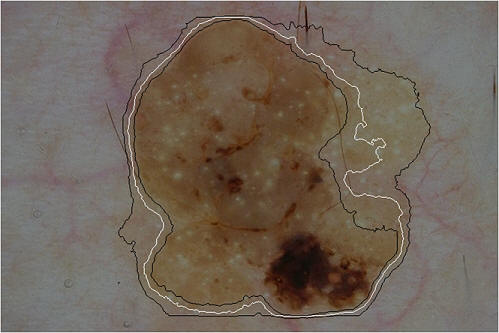
An example probabilistic segmentation of a seborrheic keratosis. White: Maximum likelihood segmentation (segmentation='lesion' if p>0.5) Black: 80% confidence intervals. More details of this technique applied to segmentation can be found in this paper.
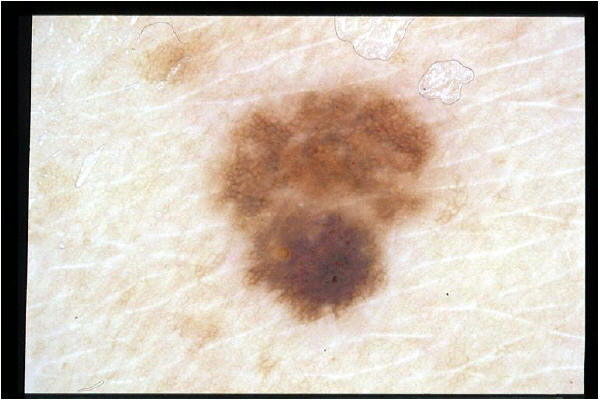
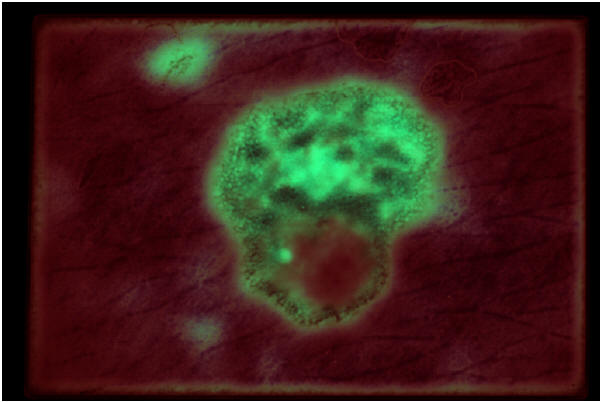
Labeling an image of a nevus for the demoscopic structure pigment network . (red: pigment_network='absent'; green: pigment_network='typical')

Labeling phantom hair. Future work will explore the use of shape priors to improve results.
